Chapter 2 Statistics for Data Science
We will skip the introductory statistics. There are many courses/ textbook out there. Recommended coursebook:
- Mean, Median, Mode, Variance, Standard Deviation
- Sample, Population
- Expected values, Estimation
- Probability
- Permutations and Combinations
- Distributions (Binomial, Normal, Poisson, Hypergeometric, etc.)
- Test Statistic and statistical inference (Z-test, T-Test, Confidence interval, etc.)
- Hypothesis Testing
R is equipped with many statistical functions. We can calculate metrics such mean, mean, median, mean, max, etc. of the given vector using functions in R.
x=rnorm(1000,100,15) #Generates a sample of size 1000 from a normal distribution with mu=100 and sigma=15
mean_x = mean(x) #Computes the sample mean.
sd_x = sd(x) #Computes the sample st dev.
sum_x = sum(x) #Computes the sum of the elements of x.
print(paste0("mean_x = ", mean_x, " sd_x = ", sd_x, " sum_x = ", sum_x))## [1] "mean_x = 100.328393193161 sd_x = 14.5746365457083 sum_x = 100328.393193161"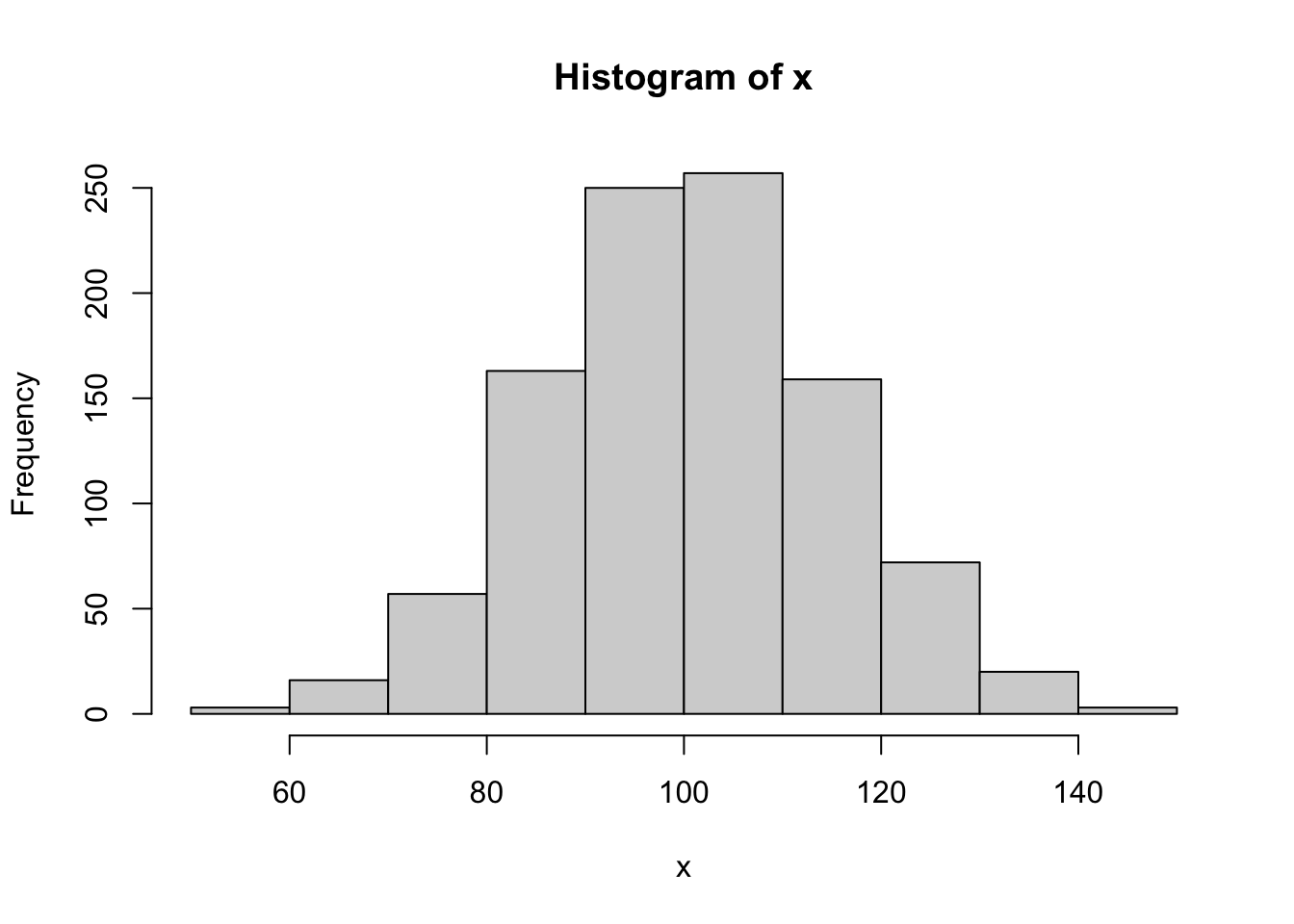
u=runif(1000,10,20) #Generates a sample of size 1000 from a uniform
#Distribution on the interval [10,20].
min_u = min(u) #Finds the smallest component of u.
max_u = max(u) #Finds the largest component of u.
sorted_u=sort(u)
length(sorted_u)## [1] 1000## [1] 10.01506## [1] 19.99784x=rnorm(1000,100,15) #Random x values.
epsilon=rnorm(1000,0,5) #Random "errors".
y=x+epsilon #Random y values dependent on x.
plot(x,y) #Scatterplot of (x,y) pairs.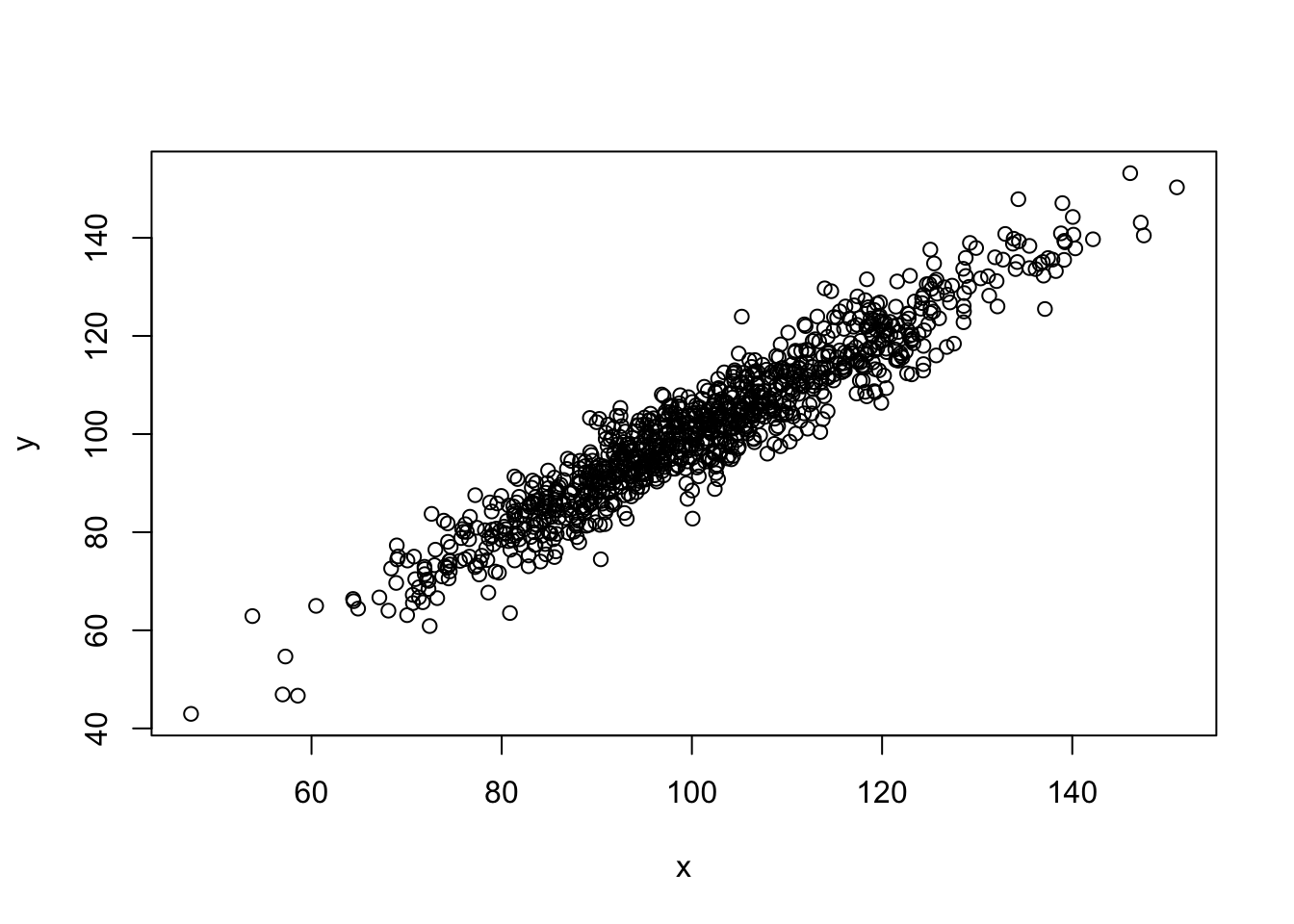
2.1 Non Parametric Statistics
Parametric statistics are the ones where we know the behavior of the population. Usually the parameters of a statistical methods are the mean and the standard deviation. Eg. confidence interval
In contrast, in non-parametrc statistics we do not need to make any assumption about the population. Non parametric methods are easy to use and apply. Refer to Conover Practical Nonparametric statistics
2.1.1 Sign Test and variations
Use to test if preferance A vs B
- Number of response with positive preferance \([num\ +]\)
- Number of response with negative preferance \([num\ -\)
- Number of response with no preferance. We do not care for those
- We test the hypothesis that:
- \(H_0: P(+) = P(-)\ vs\ H_1: P(+) ! = P(_)\)
- with \(n = [num \ +] + [num \ -]\)
- and test statistic \(T = [num \ +]\)
- Null Distribution \(T \~ B(n, 0.5)\)
McNemar Test of Significant Changes (sign test variant)
Eg: differenct between voter intentions before and after debate
Cox-Stuart Test for Trend (sign test variant)
Eg. Given the temperature for last 19 January Check if trend in temperature pattern exists. (hint: convert the temp between two consecutive January to + or - based on pattern and count those up)
2.1.2 chi squared tests for contengency tables
2.1.3 Median test
To test whether the sample median is different from grand median (population median)
2.1.4 Measure of Dependence
2.1.5 Chi-Squared goodness of fit tests
2.1.7 Mann - Whitney - Wilcoxin Test
Tests if two samples are independent.
- Assumes that two samples are from different population.
- Assumes that samples are statistically independent of each other
- Measurement scale is at least ordinal
- Null Hypothesis: Two probability distributions are identitcal
- Rank observations from smallest to largest
- Test Statistic \(T = \sum _{i=1}^{n} Rank(X_i)\)
Steps:
- sort the values
- Assign Rank
- Replace Ties
- Calcualte T
- Calculate W
#Example: Farm Boys and Town Boys
#Creating data set
residence=c(rep("Farm",12),rep("Town",36))
residence=as.factor(residence)
fitness=c(c(14.8,7.3,5.6,6.3,9.0,4.2),
c(10.6,12.5,12.9,16.1,11.4,2.7),
c(12.7,14.2,12.6,2.1,17.7,11.8),
c(16.9,7.9,16.0,10.6,5.6,5.6),
c(7.6,11.3,8.3,6.7,3.6,1.0),
c(2.4,6.4,9.1,6.7,18.6,3.2),
c(6.2,6.1,15.3,10.6,1.8,5.9),
c(9.9,10.6,14.8,5.0,2.6,4.0))
mydata=data.frame(residence,fitness)
#Mann-Whitney-Wilcoxon Test
wilcox.test(fitness~residence,data=mydata)## Warning in wilcox.test.default(x = c(14.8, 7.3, 5.6, 6.3, 9, 4.2, 10.6, : cannot compute exact p-
## value with ties##
## Wilcoxon rank sum test with continuity correction
##
## data: fitness by residence
## W = 243, p-value = 0.5279
## alternative hypothesis: true location shift is not equal to 0Calculating Wilcoxin (mann-whitney-wilcoxin) Test
## [1] 48 2mydata=cbind(mydata,rank=1:48)
#Replacing ties
mydata$rank[12:14]=13
mydata$rank[20:21]=20.5
mydata$rank[29:32]=30.5
mydata$rank[41:42]=41.5
#Calculating T
T_value = sum(mydata$rank[mydata$residence=="Farm"])
T_value## [1] 321## [1] 2432.1.8 Kruskal-Wallis Test
Use it for Several Independent Samples. Extends mann-whitney-wilcoxin test for more than two samples using same assumptions and hypothesis
#Example: Corn Yields
corn.yield=c(c(83,91,94,89,89,96,91,92,90),
c(91,90,81,83,84,83,88,91,89,84),
c(101,100,91,93,96,95,94),
c(78,82,81,77,79,81,80,81))
farming.method=as.factor(c(rep(1,9),
rep(2,10),
rep(3,7),
rep(4,8)))
mydata2=data.frame(corn.yield,farming.method)
#Plot of data
plot(farming.method,corn.yield,data=mydata2)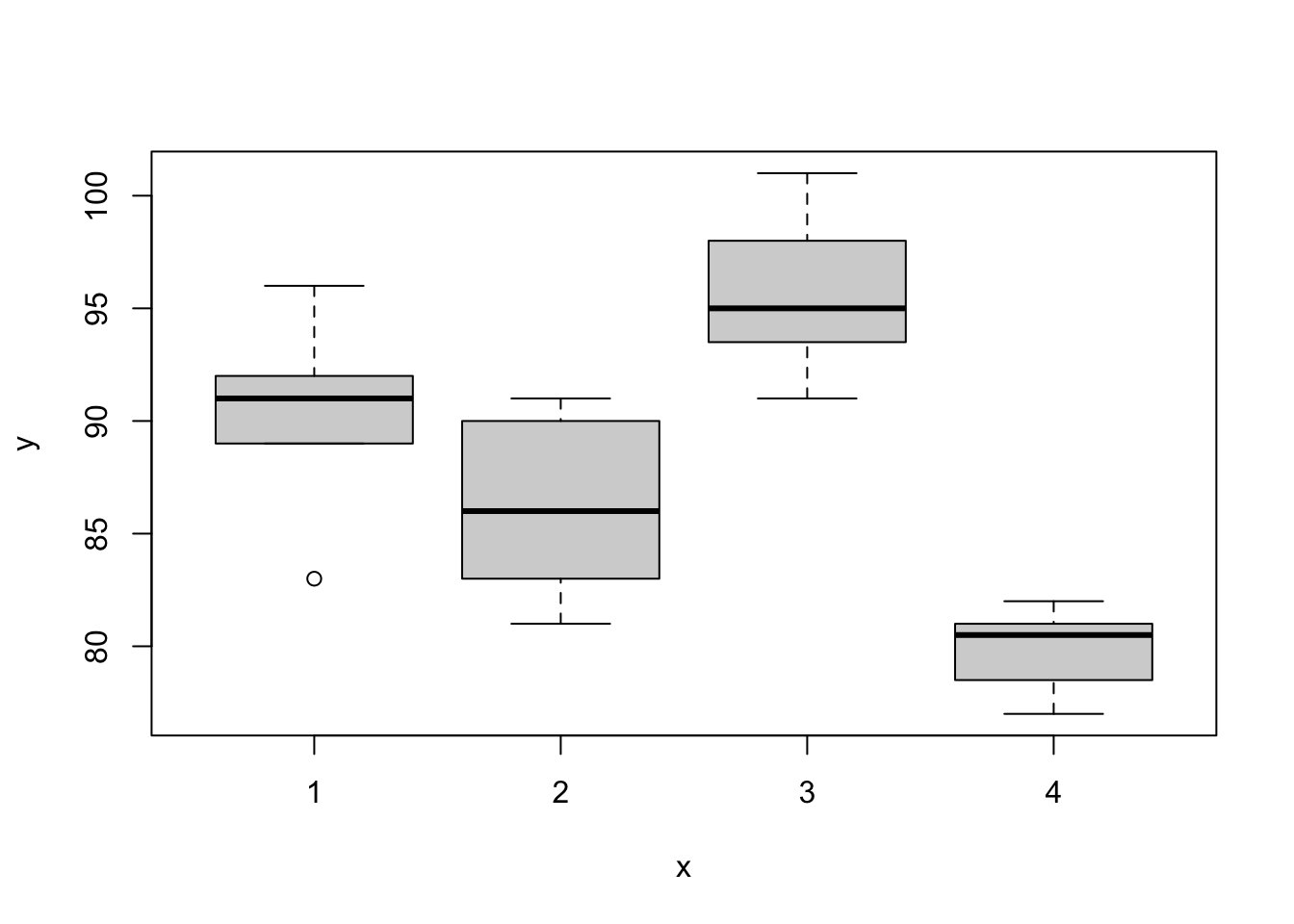
##
## Kruskal-Wallis rank sum test
##
## data: corn.yield by farming.method
## Kruskal-Wallis chi-squared = 25.629, df = 3, p-value = 1.141e-05##
## Kruskal-Wallis rank sum test
##
## data: fitness by residence
## Kruskal-Wallis chi-squared = 0.41362, df = 1, p-value = 0.52012.1.9 Wilcoxin-signed-rank Test
For matched pairs
- Random sample of pairs
- Null Hypothesis that exepcted value of two samples are same
- Discard ties and calculate absolute difference (since difference between the pairs is 0)
Steps:
- Calculate differences
- Discard ties
- Sort data based on absolute differences
- Create Rank
- Replace ties
- Create Signed Ranks
- Calculate R
- Calculate Tplus
- Run mann-whitney-wilcox test
#Example: Twins
firstborn=c(86,71,77,68,91,72,77,91,70,71,88,87)
secondborn=c(88,77,76,64,96,72,65,90,65,80,81,72)
#Wilcox Signed Ranks Test
wilcox.test(secondborn,firstborn,paired=TRUE)## Warning in wilcox.test.default(secondborn, firstborn, paired = TRUE): cannot compute exact p-value
## with ties## Warning in wilcox.test.default(secondborn, firstborn, paired = TRUE): cannot compute exact p-value
## with zeroes##
## Wilcoxon signed rank test with continuity correction
##
## data: secondborn and firstborn
## V = 24.5, p-value = 0.4765
## alternative hypothesis: true location shift is not equal to 0Step by step exapmple
#Calculating differences
mydata3=data.frame(firstborn,
secondborn,
D=secondborn-firstborn,
absD=abs(secondborn-firstborn))
#Discarding ties
mydata3=mydata3[mydata3$D!=0,]
#Sort data based on absD
mydata3=mydata3[order(mydata3$absD),]
#Creating absD ranks
mydata3=cbind(mydata3,absD.rank=1:11)
#Replacing absD rank ties
mydata3$absD.rank[1:2]=1.5
mydata3$absD.rank[5:6]=5.5
#Creating signed ranks R
R=(mydata3$absD.rank)*sign(mydata3$D)
mydata3=cbind(mydata3,R)
#Calculating Tplus
Tplus=sum(R[R>0])
Tplus## [1] 24.5## Warning in wilcox.test.default(secondborn, firstborn, paired = TRUE): cannot compute exact p-value
## with ties## Warning in wilcox.test.default(secondborn, firstborn, paired = TRUE): cannot compute exact p-value
## with zeroes##
## Wilcoxon signed rank test with continuity correction
##
## data: secondborn and firstborn
## V = 24.5, p-value = 0.4765
## alternative hypothesis: true location shift is not equal to 0## Warning in wilcox.test.default(firstborn, secondborn, paired = TRUE): cannot compute exact p-value
## with ties## Warning in wilcox.test.default(firstborn, secondborn, paired = TRUE): cannot compute exact p-value
## with zeroes##
## Wilcoxon signed rank test with continuity correction
##
## data: firstborn and secondborn
## V = 41.5, p-value = 0.4765
## alternative hypothesis: true location shift is not equal to 0## [1] -41.52.1.10 Quade Test
For Several Related Samples
#Example: Hand Lotion Sales
y <- matrix(c( 5, 4, 7, 10, 12,
1, 3, 1, 0, 2,
16, 12, 22, 22, 35,
5, 4, 3, 5, 4,
10, 9, 7, 13, 10,
19, 18, 28, 37, 58,
10, 7, 6, 8, 7),
nrow = 7, byrow = TRUE,
dimnames =
list(Store = as.character(1:7),
Brand = LETTERS[1:5]))
y## Brand
## Store A B C D E
## 1 5 4 7 10 12
## 2 1 3 1 0 2
## 3 16 12 22 22 35
## 4 5 4 3 5 4
## 5 10 9 7 13 10
## 6 19 18 28 37 58
## 7 10 7 6 8 7##
## Quade test
##
## data: y
## Quade F = 3.8293, num df = 4, denom df = 24, p-value = 0.015192.1.11 Spearman’s Test
Measures of rank correlation
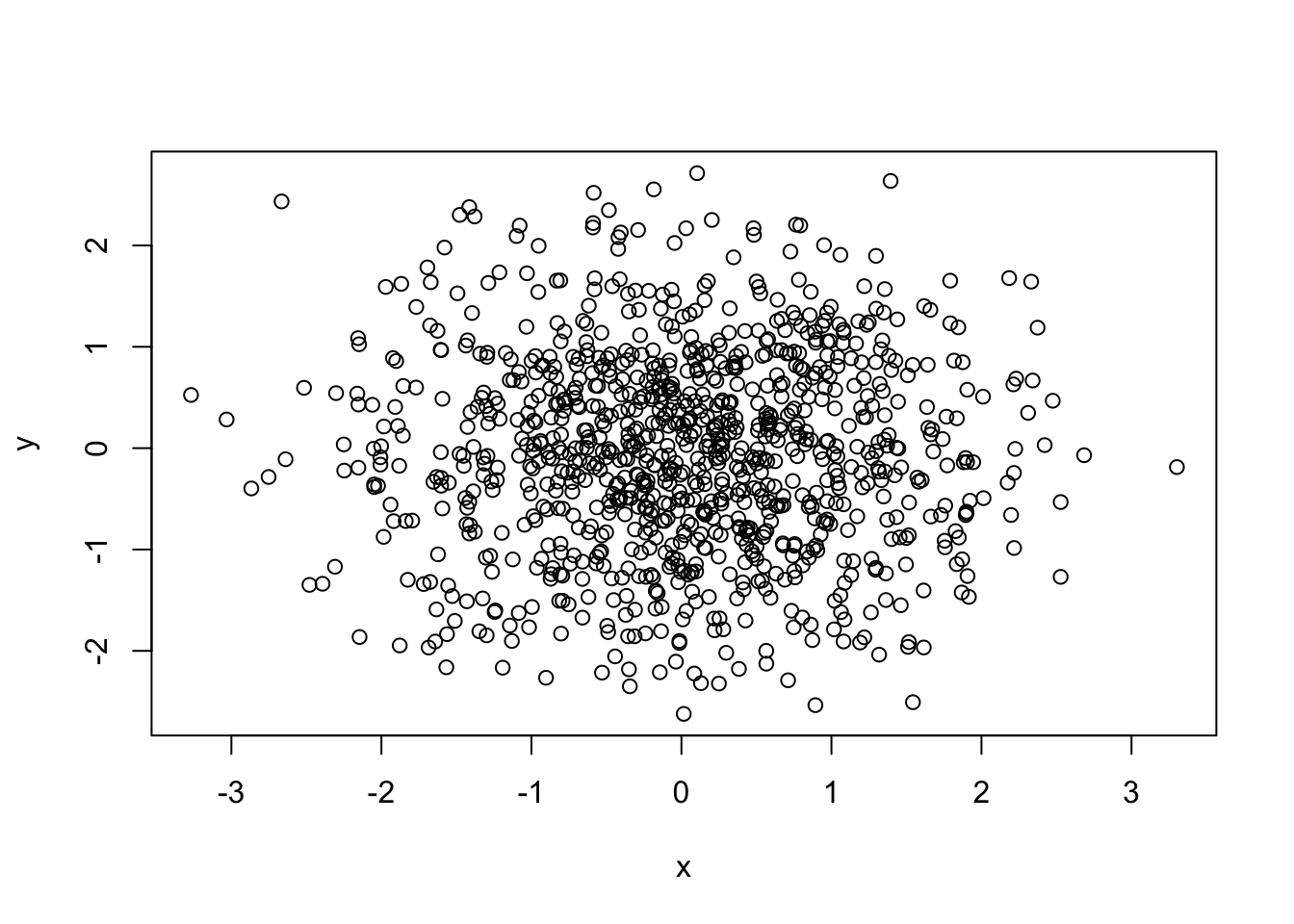
## [1] -0.005149681## [1] 0.0004317244##
## Pearson's product-moment correlation
##
## data: x and y
## t = -0.16269, df = 998, p-value = 0.8708
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.06712134 0.05686155
## sample estimates:
## cor
## -0.005149681##
## Spearman's rank correlation rho
##
## data: x and y
## S = 166594546, p-value = 0.9891
## alternative hypothesis: true rho is not equal to 0
## sample estimates:
## rho
## 0.0004317244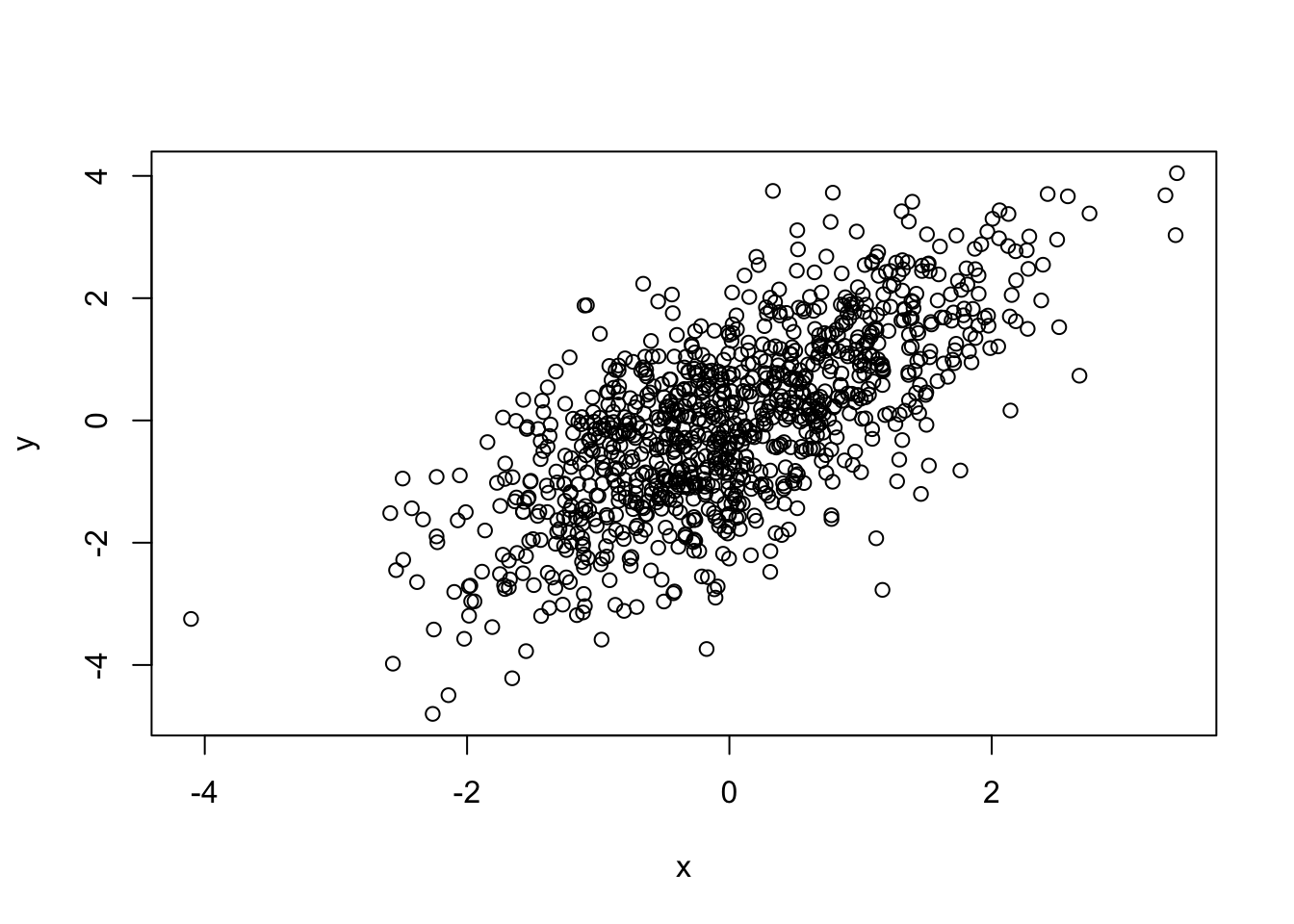
## [1] 0.6901655## [1] 0.6645341##
## Pearson's product-moment correlation
##
## data: x and y
## t = 30.129, df = 998, p-value < 2.2e-16
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.6562504 0.7212975
## sample estimates:
## cor
## 0.6901655##
## Spearman's rank correlation rho
##
## data: x and y
## S = 55910934, p-value < 2.2e-16
## alternative hypothesis: true rho is not equal to 0
## sample estimates:
## rho
## 0.66453412.1.12 Assessing Normality
Using quantile-quantile-plots and Hypothesis Tests
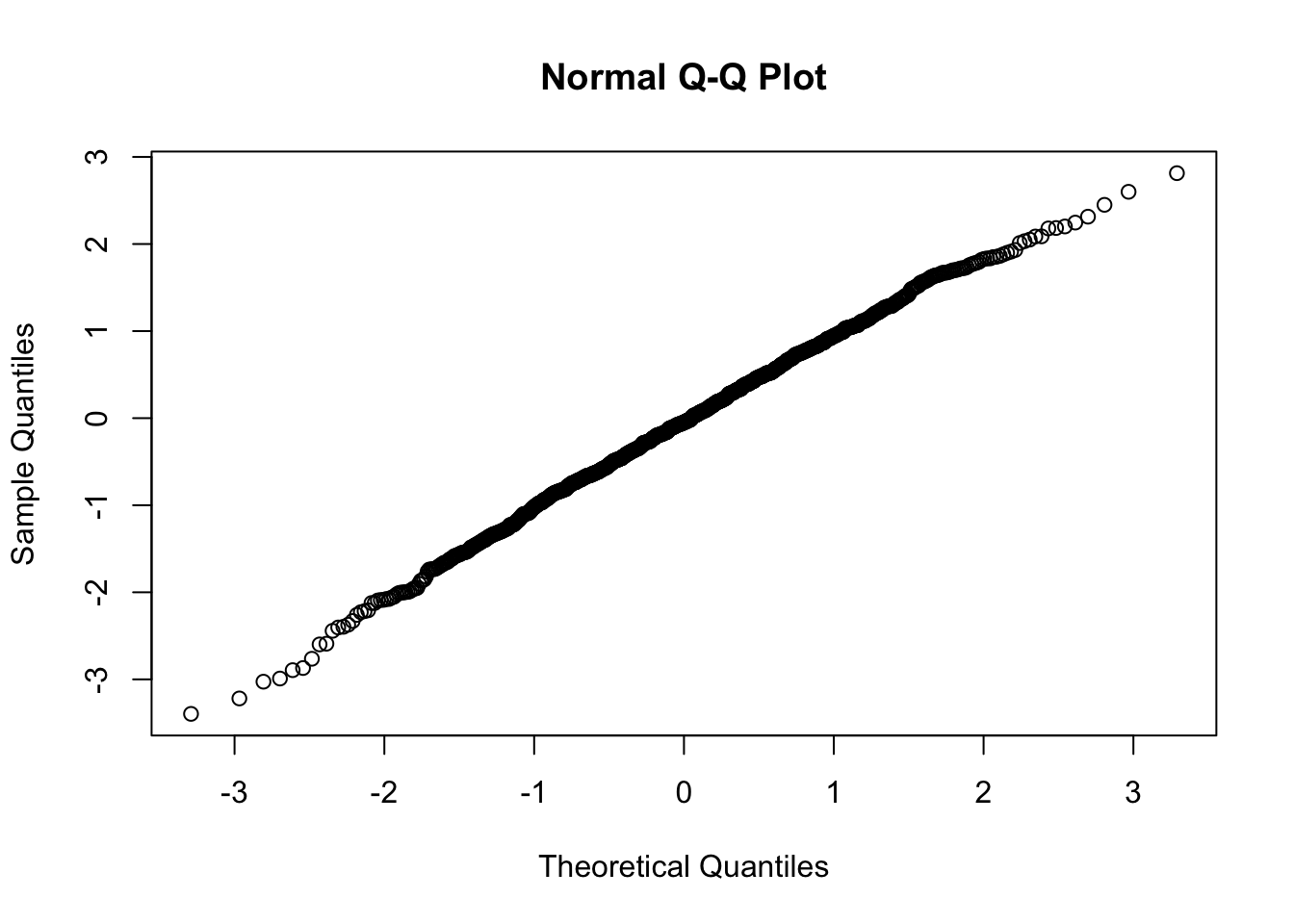
Using Kolmogorov-Smirnov Test
##
## One-sample Kolmogorov-Smirnov test
##
## data: x
## D = 0.024585, p-value = 0.5812
## alternative hypothesis: two-sidedUsing Shapiro-Wilk Test.
##
## Shapiro-Wilk normality test
##
## data: x
## W = 0.99753, p-value = 0.1367Normality example using all methods explained
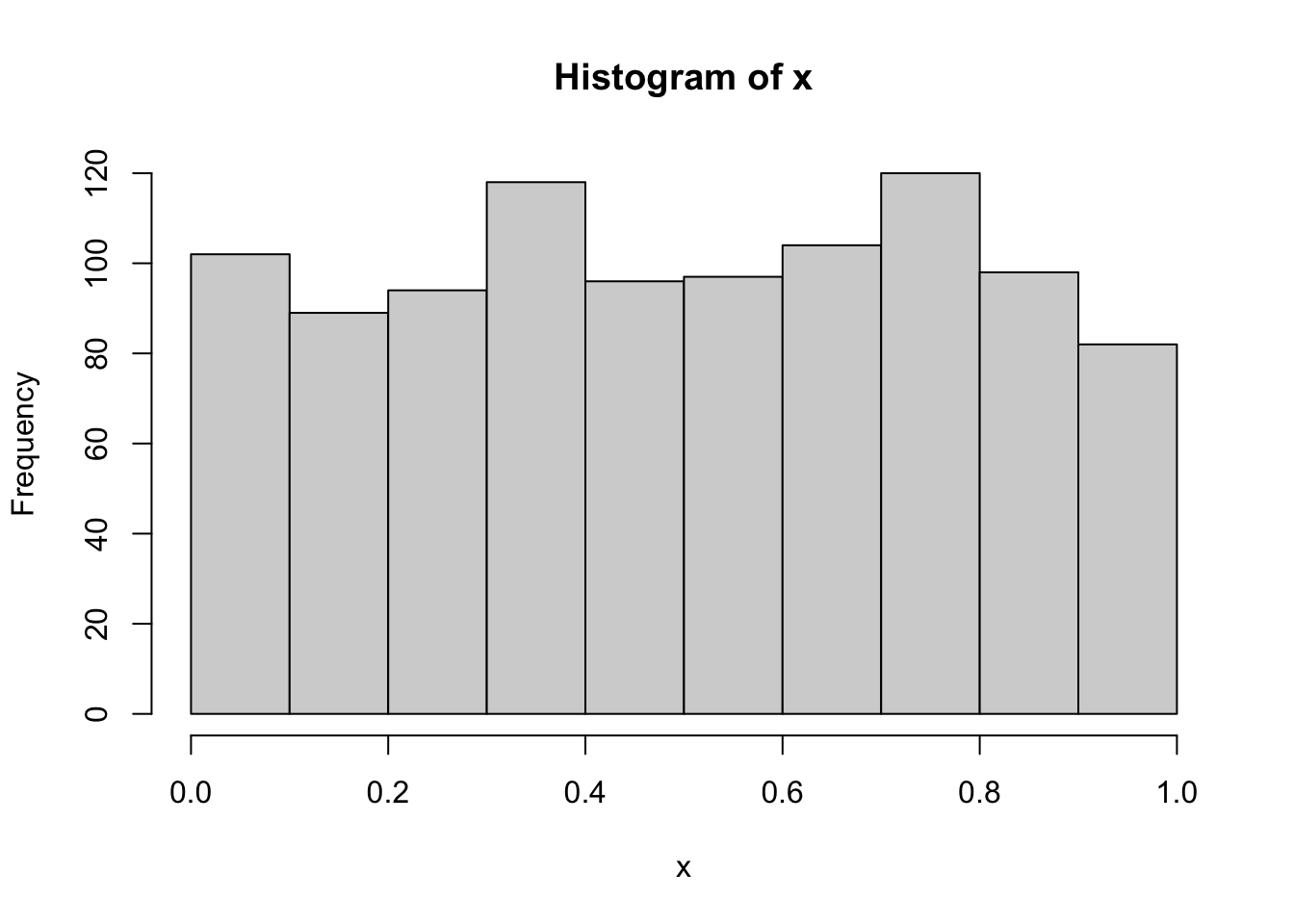
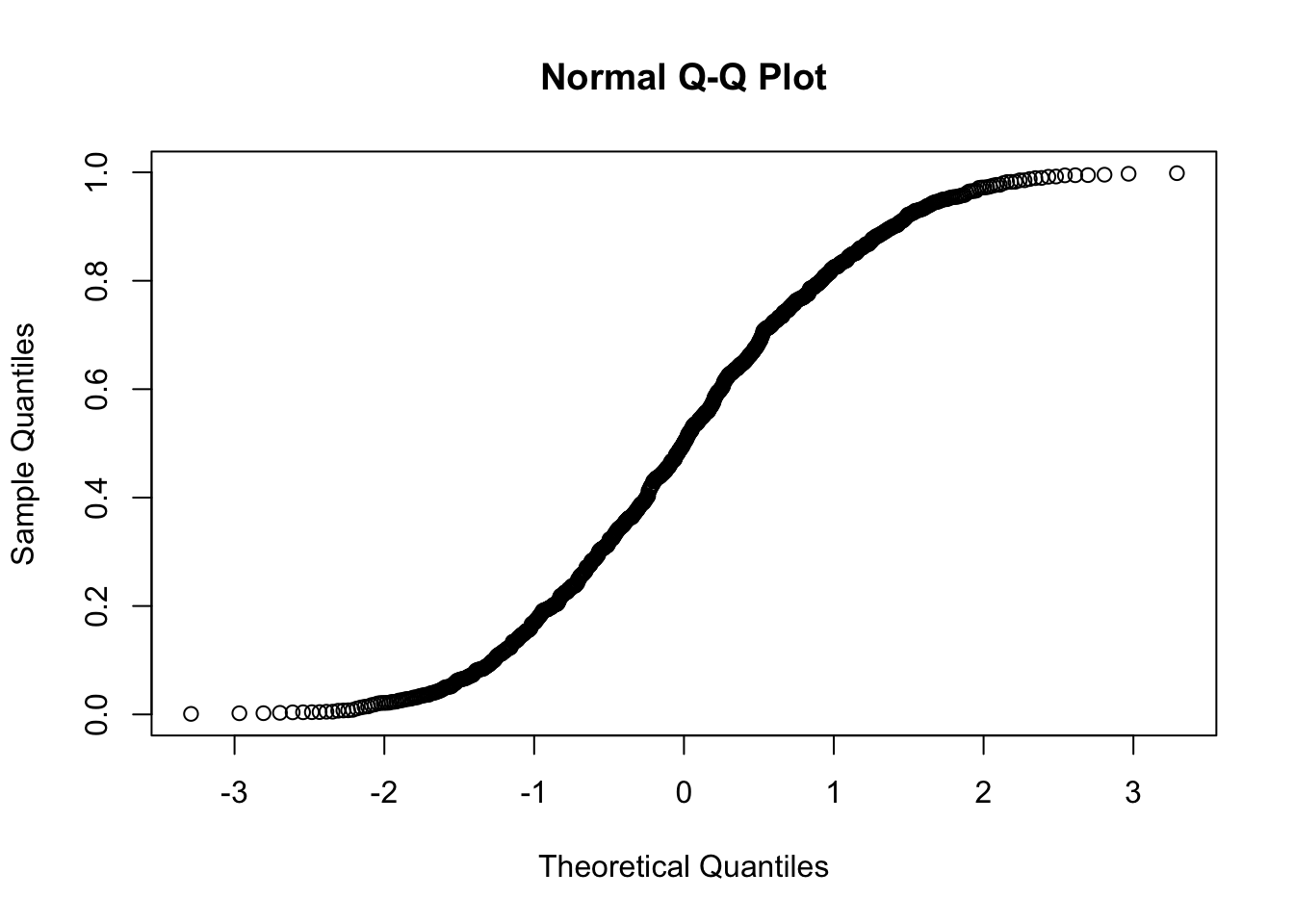
#Kolmogorov-Smirnov Test
#Null hypothesis is that x and y have the same distribution
ks.test(x,y="pnorm")##
## One-sample Kolmogorov-Smirnov test
##
## data: x
## D = 0.5004, p-value < 2.2e-16
## alternative hypothesis: two-sided##
## One-sample Kolmogorov-Smirnov test
##
## data: x
## D = 0.024197, p-value = 0.6017
## alternative hypothesis: two-sided##
## Shapiro-Wilk normality test
##
## data: x
## W = 0.95951, p-value = 5.12e-16