Chapter 4 Linear Regression
Concepts:
- Regression Line
- Correlation
- Mean Squared Errors and Root Mean Squared Errors
- Estimate \(\hat{\sigma}\)
- Ordinary Least Squares (OLS) Estimator \(\hat{\beta}\) BLUE
- Covariance matrix \(\widehat{cov}(\hat{\beta})\)
- Standard Error
- Model output (t-values, p-values, F-statistics, p-value of F-statistic)
- OLS Assumptions
4.1 Simple Linear Regression
Lets look at the Old faithful eruptions data again. In the dataset have eruption duration (mins) and waiting time between each eruption. We can look at the scatterplot and the line that best describes the pattern
| eruptions | waiting |
|---|---|
| 3.600 | 79 |
| 1.800 | 54 |
| 3.333 | 74 |
| 2.283 | 62 |
| 4.533 | 85 |
| 2.883 | 55 |
ggplot(faithful, aes(x= waiting, y = eruptions)) +
geom_point() +
geom_smooth(method = 'lm', formula = y~x)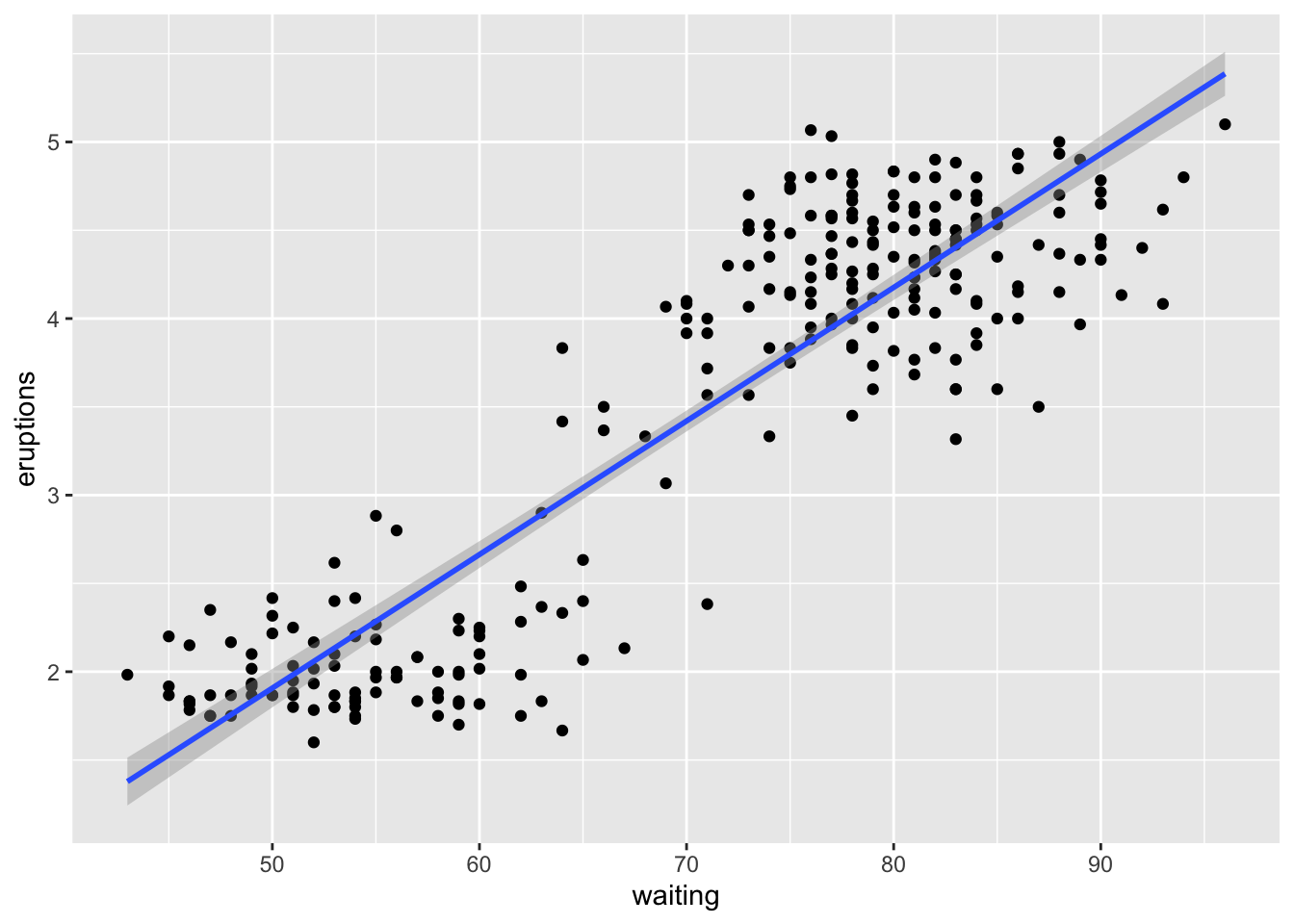
Here we can see that the length of the eruptions are “positively correlated” with the waiting time i.e. longer the waiting time, longer the eruptions. We can also see that the points are clustered around two areas. The line that best fits the data is known as regression line.
We can do some basic calculations to build this linear model using r.
Mean Eruption times = 3.4877831
Variance Eruption times = 1.3027283
Standard Deviation of Eruption times = 1.1413713
Mean wait times = 70.8970588
Variance wait times = 184.8233124
Standard Deviation of wait times = 13.5949738
Correlation coefficient = 0.8974994
Slope of regression line =
Mean Square Errors
Running inbuilt function to run linear regression
##
## Call:
## lm(formula = eruptions ~ waiting, data = faithful)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.29917 -0.37689 0.03508 0.34909 1.19329
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.874016 0.160143 -11.70 <2e-16 ***
## waiting 0.075628 0.002219 34.09 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.4965 on 270 degrees of freedom
## Multiple R-squared: 0.8115, Adjusted R-squared: 0.8108
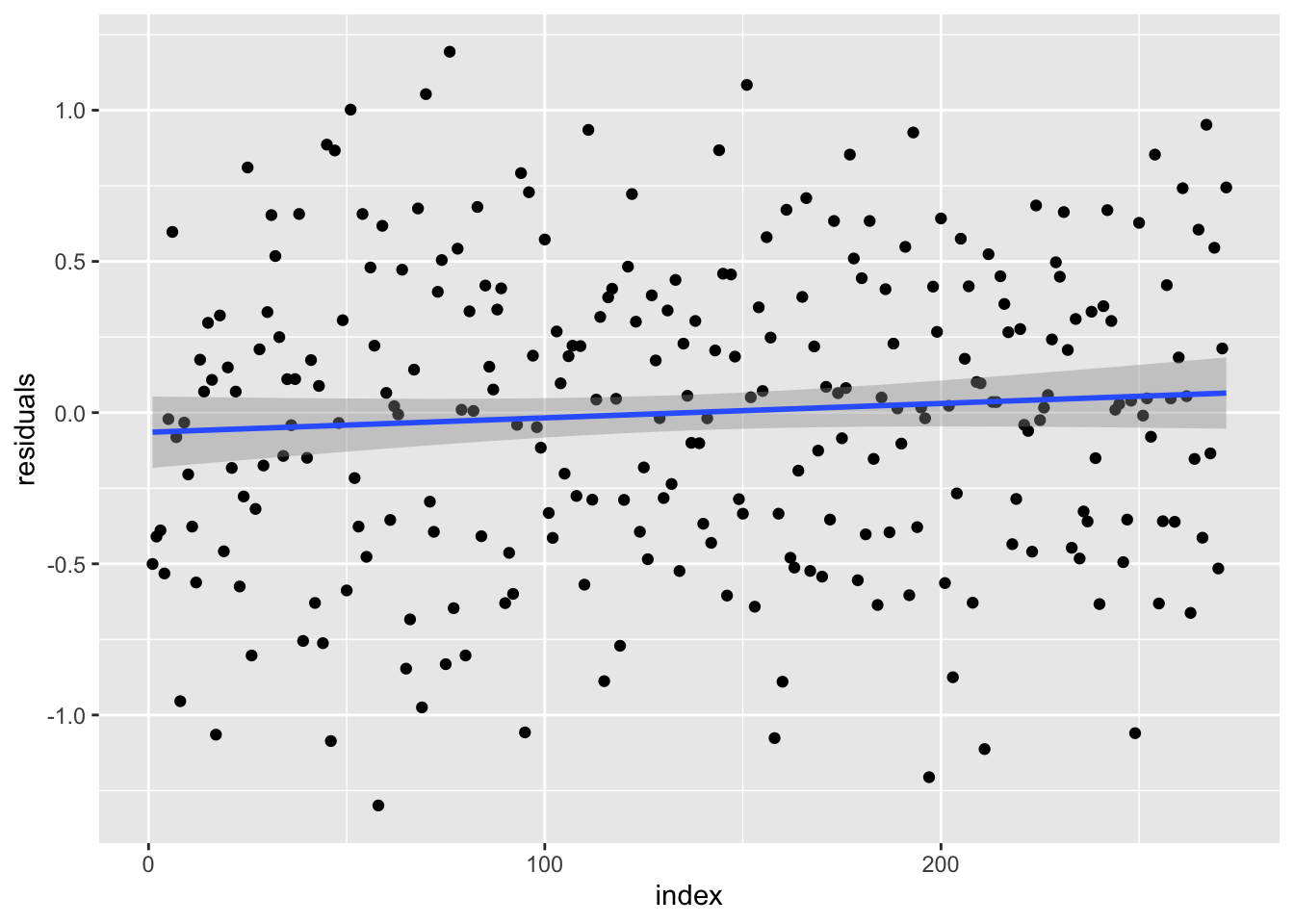
## F-statistic: 1162 on 1 and 270 DF, p-value: < 2.2e-16Explanation of the model here {residuals, coefficients, Standard-errors (including residuals), significance, F-statistic}
Residual plot
4.2 Multiple Linear Regression
Extend simple linear regression to multiple linear regression.
Functional form \[y_i = \beta_1x_{i1} + ... \beta_px_{ip} + \epsilon_i \] We do not know true beta
\[Y = X\beta + \epsilon\] So we use \(\hat\beta\) to estimate \(\beta\) and OLS gives us the best result. Sum of squred of errors is minimized when \(\hat\beta = (X'X)^{-1}X'Y\)
Residuals \(e= Y - X\hat\beta\) and e is orthogonal to X
Ordinary Least Squares (involves some Matrix Algebra and mathematical proofs). Assumptions:
- Design matrix X has full rank. ie. there are more rows of data than the number of independent variables (features) in the dastaset
- The unobservable random vector \(\epsilon\) is Independent and Identically distributed (IID) with mean 0 and variance of more than 0.
- If design matrix X is random then \(\epsilon\) is independent of X.
Clarify the the difference between beta/betahat and e/epsilon
When the disturbance terms epsilon are normally distributed, the beta hat are the Maximum likely estimator for beta
Gradient Decent method Elaborate later
Define Loss Function Loss = residual . Calculate the Mean Squared of Errors
\[ E = \frac 1 n \sum_{i=0}^n(y_i - \bar y_i)^2 = \frac 1 n \sum_{i=0}^n(y_i - (mx_i +c))^2 \] Calculate the Gradient ie. partial derivative with respect to m and c to find the constants where the \(E\) is the smallest. Which are
\(D_m = \delta_m/\delta_y = \frac {-2} n \sum_{i=0}^n x_i(y_i - \bar y_i)\)
\(D_c = \delta_c/\delta_y = \frac {-2} n \sum_{i=0}^n (y_i - \bar y_i)\)
Update \(m\) and \(c\) using \(m = m - L \times D_m\) and \(c=c-L \times D_c\)
Repeat the \(m\) and \(c\) caluclation, plug the value into Loss function with value of x and y. The required \(m\) and \(c\) are when \(E = 0\).
Also need to avoid the local minima values so use multiple random starting point for values of \(m\) and \(c\) and do calculation multiple times (epochs)
Few things to test to ensure model fit
- plot residual vs x (should have no trend)
- Mean of residul should center around zero
- Assumption that residual are normally distributed
4.2.1 Consequences of Incorrect Functional Form
Creating data
set.seed(5366)
x=1:100
X=cbind(1,x,x^2)
beta=c(10,-1.15,0.0205)
y=X%*%beta+5*rnorm(100)
mydata=data.frame(y,x)
#Plot
plot(y~x,data=mydata)
#Fitting Quadratic Model
model2=lm(y~x+I(x^2),data=mydata)
summary(model2)##
## Call:
## lm(formula = y ~ x + I(x^2), data = mydata)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.6521 -3.2214 -0.1858 2.9871 11.4882
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 11.453364 1.428003 8.021 2.43e-12 ***
## x -1.208440 0.065264 -18.516 < 2e-16 ***
## I(x^2) 0.020854 0.000626 33.311 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.665 on 97 degrees of freedom
## Multiple R-squared: 0.9774, Adjusted R-squared: 0.9769
## F-statistic: 2098 on 2 and 97 DF, p-value: < 2.2e-16plot(y~x,data=mydata)
x.plot.vector=1:100
y.plot.vector=predict(model2,
newdata=data.frame(x=x.plot.vector))
lines(x.plot.vector,
y.plot.vector,
col="red")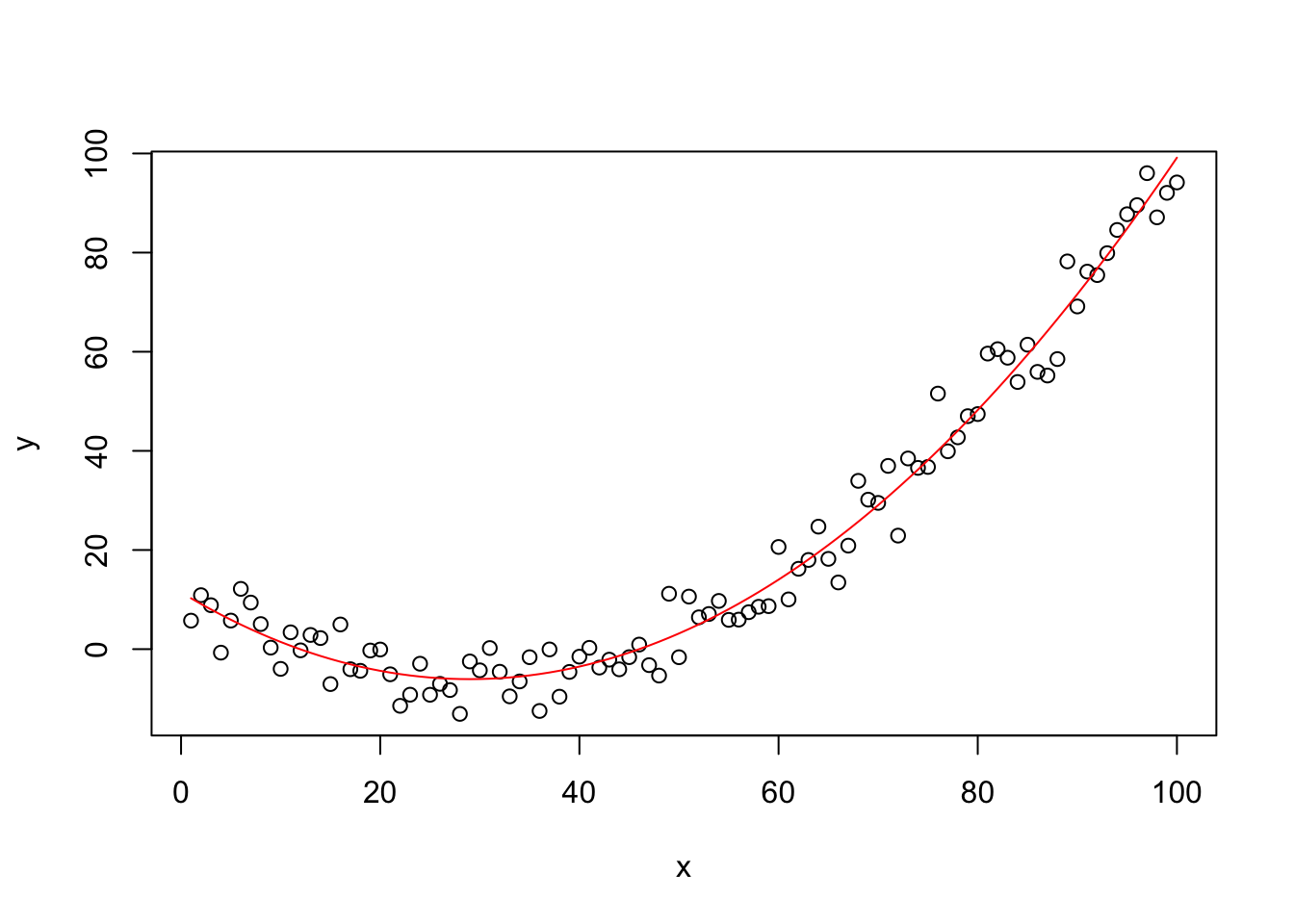
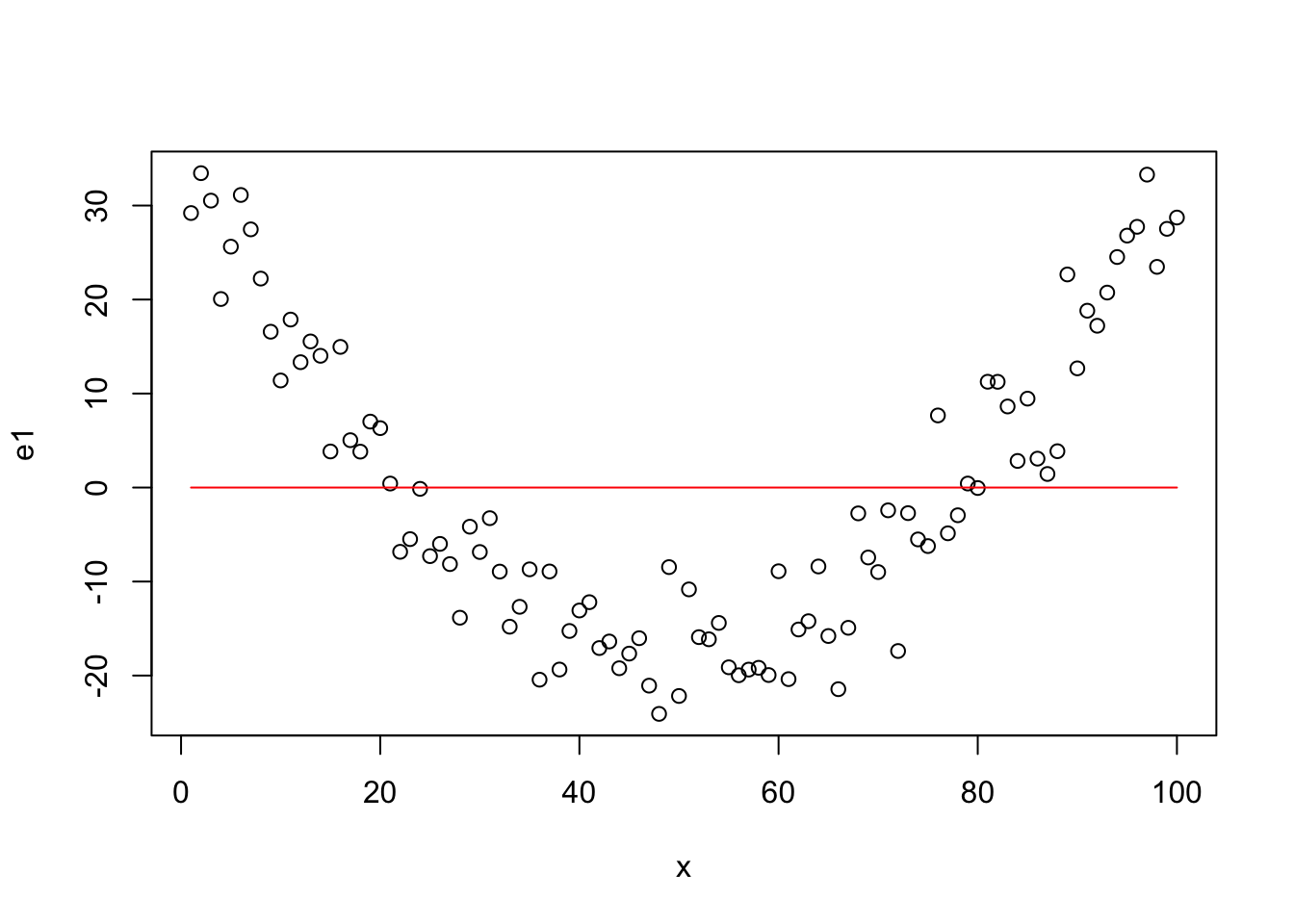
Residuals for Quadratic Model
betahat2=coef(model2)
X2=model.matrix(model2)
e2=y-X2%*%betahat2
#e2
e2=residuals(model2)
#e2
plot(x,e2)
lines(x,rep(0,length(x)),col="red")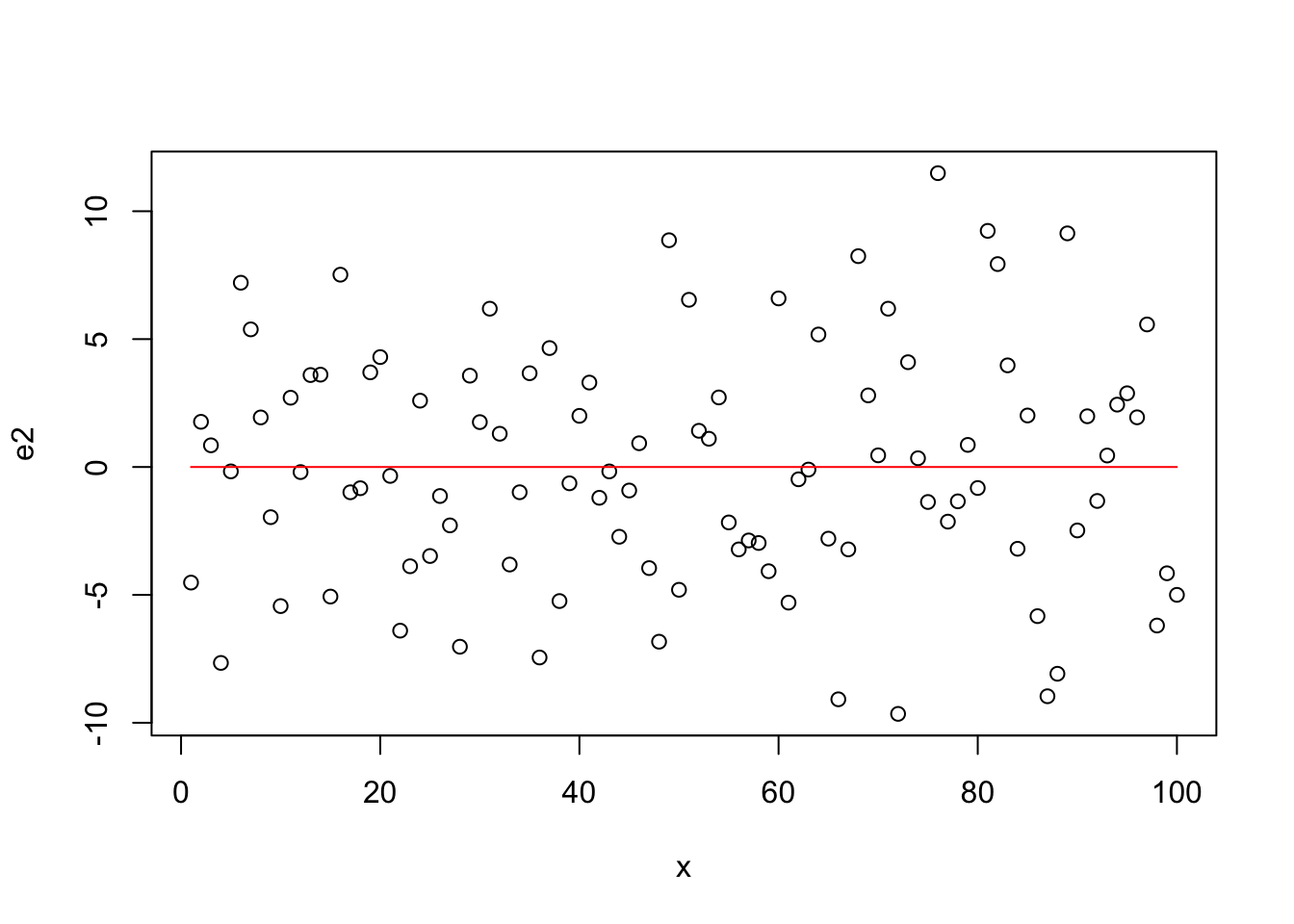
Fitting a linear model
##
## Call:
## lm(formula = y ~ x, data = mydata)
##
## Residuals:
## Min 1Q Median 3Q Max
## -24.077 -14.502 -3.712 12.844 33.444
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -24.35330 3.29860 -7.383 5.13e-11 ***
## x 0.89783 0.05671 15.833 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 16.37 on 98 degrees of freedom
## Multiple R-squared: 0.7189, Adjusted R-squared: 0.7161
## F-statistic: 250.7 on 1 and 98 DF, p-value: < 2.2e-16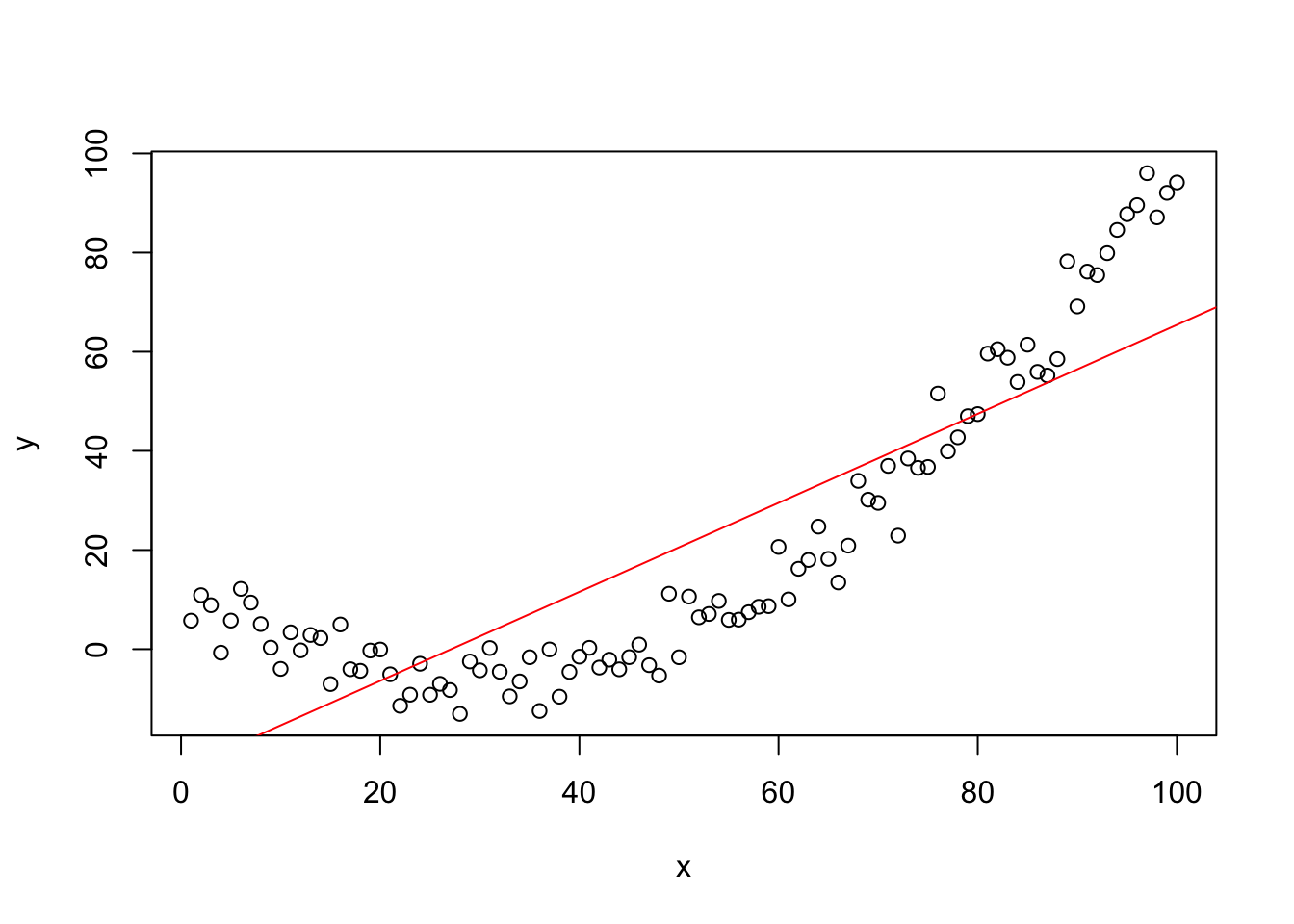
f-test Function
ftest=function(model0,model){
d0=length(coef(model0))
d=length(coef(model))
n=length(residuals(model))
F=((deviance(model0)-deviance(model))/(d-d0))/
((deviance(model))/(n-d))
return(pf(F,d-d0,n-d,lower.tail=FALSE))
}
ftest(model1,model2)## [1] 6.724646e-55##
## Call:
## lm(formula = y ~ x + I(x^2), data = mydata)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.6521 -3.2214 -0.1858 2.9871 11.4882
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 11.453364 1.428003 8.021 2.43e-12 ***
## x -1.208440 0.065264 -18.516 < 2e-16 ***
## I(x^2) 0.020854 0.000626 33.311 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.665 on 97 degrees of freedom
## Multiple R-squared: 0.9774, Adjusted R-squared: 0.9769
## F-statistic: 2098 on 2 and 97 DF, p-value: < 2.2e-16Plots involving Yhat for Quadratic Model
X2 = model.matrix(model2)
betahat2=coef(model2)
yhat2=X2%*%betahat2
plot(yhat2,y)
lines(1:100,1:100,col="red")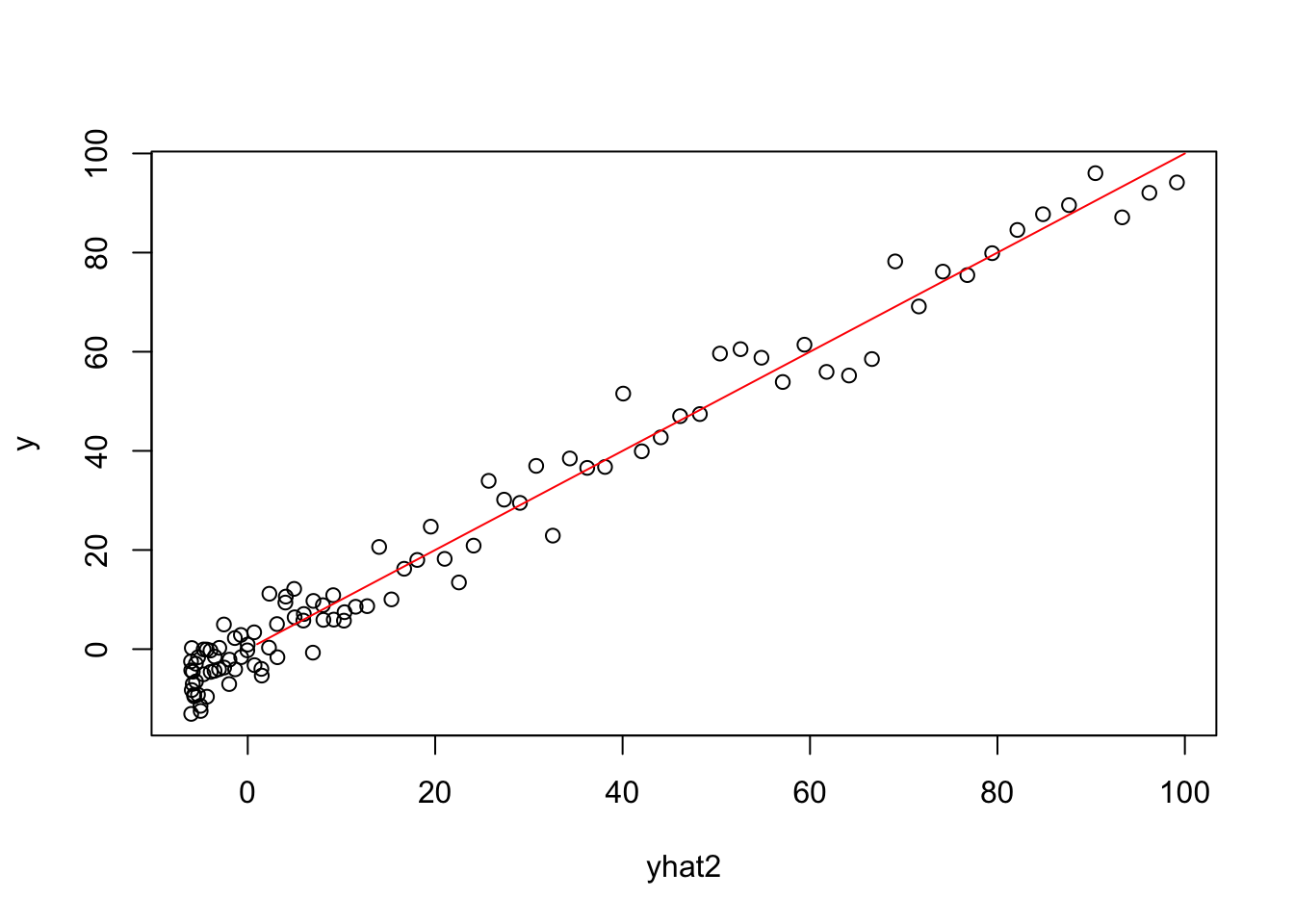
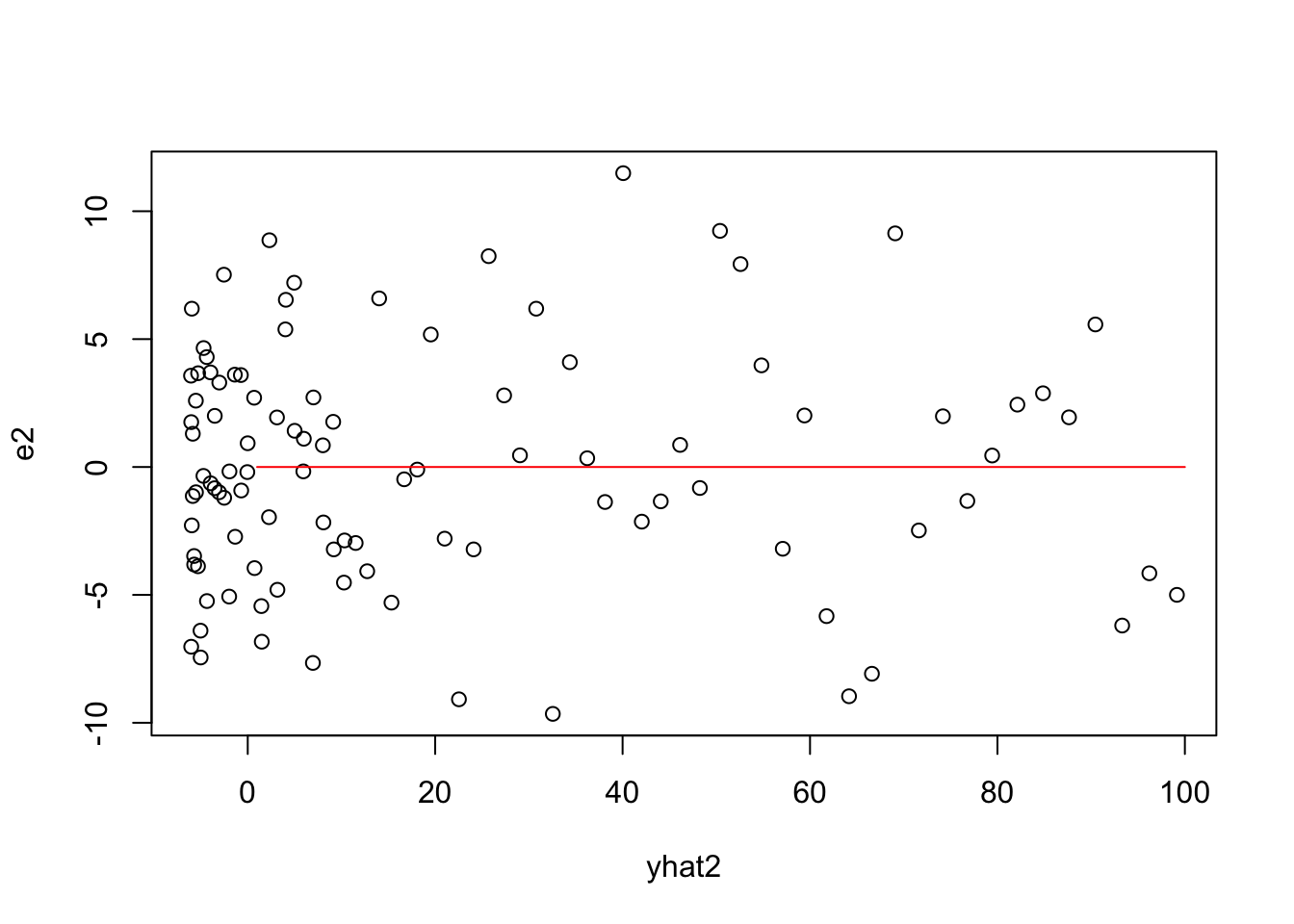
Plots involving Yhat for Linear Model
X1 = model.matrix(model1)
betahat1=coef(model1)
yhat1=X1%*%betahat1
plot(yhat1,y)
lines(1:100,1:100,col="red")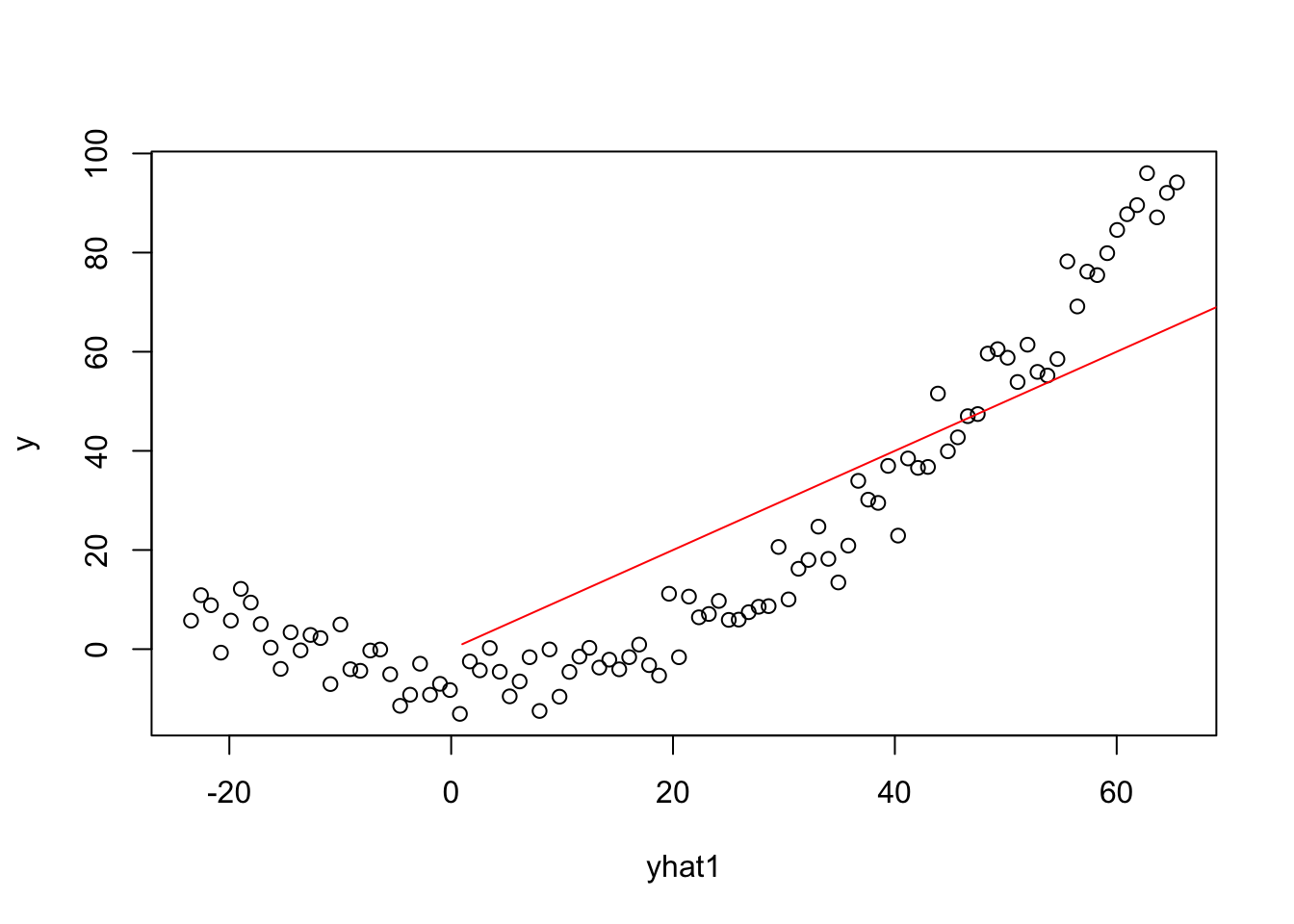
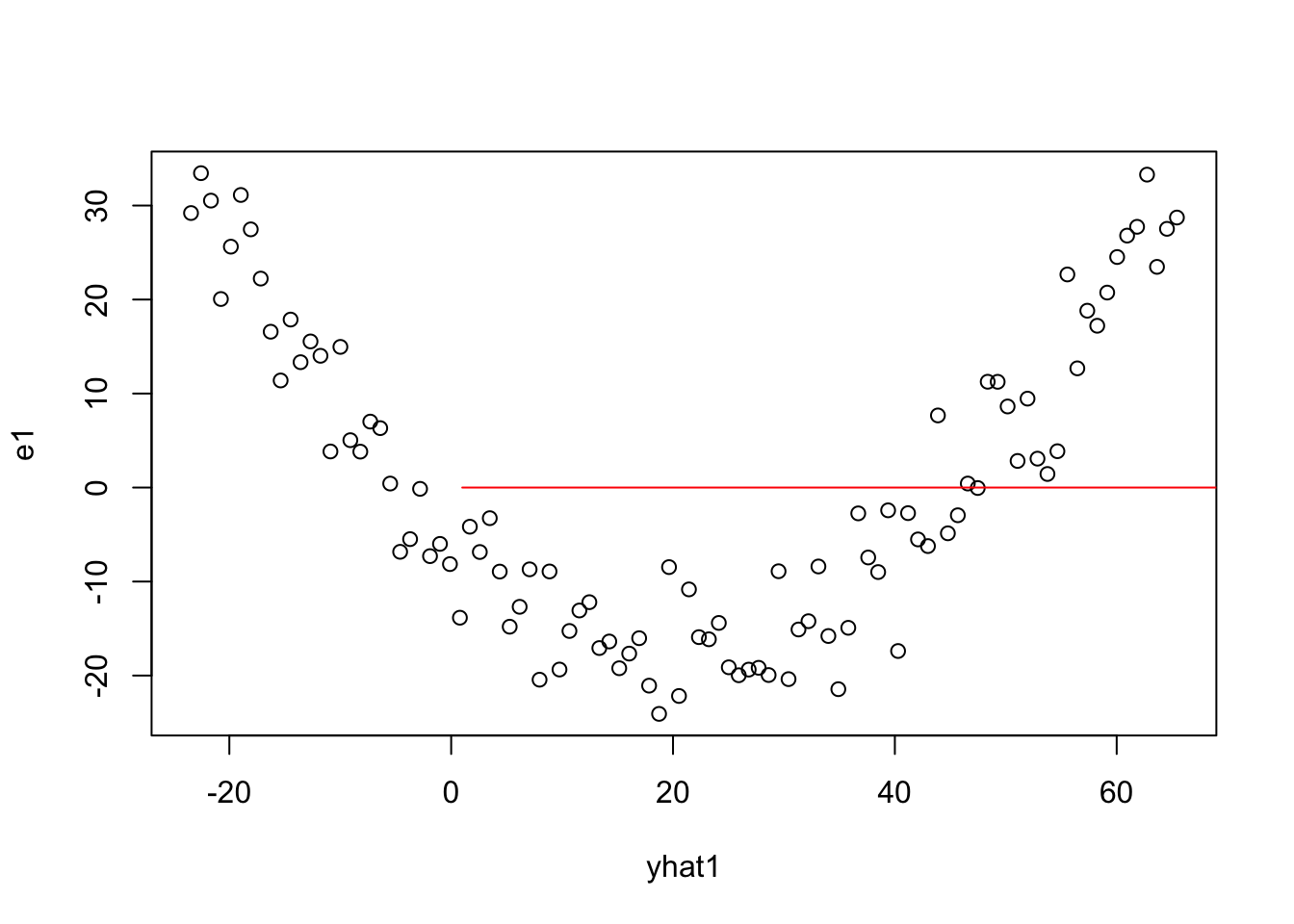
4.2.2 Design Matrix Assumptions
Rank of Design Matrix for Model2
## (Intercept) x I(x^2)
## 1 1 1 1
## 2 1 2 4
## 3 1 3 9
## 4 1 4 16
## 5 1 5 25
## 6 1 6 36## [1] 100 3## [1] 3
## attr(,"method")
## [1] "tolNorm2"
## attr(,"useGrad")
## [1] FALSE
## attr(,"tol")
## [1] 2.220446e-14## (Intercept) x I(x^2) z
## 1 1 1 1 1
## 2 1 2 4 2
## 3 1 3 9 3
## 4 1 4 16 4
## 5 1 5 25 5
## 6 1 6 36 6## [1] 100 4## [1] 3
## attr(,"method")
## [1] "tolNorm2"
## attr(,"useGrad")
## [1] FALSE
## attr(,"tol")
## [1] 2.220446e-14##
## Call:
## lm(formula = y ~ x + I(x^2) + z)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.6521 -3.2214 -0.1858 2.9871 11.4882
##
## Coefficients: (1 not defined because of singularities)
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 11.453364 1.428003 8.021 2.43e-12 ***
## x -1.208440 0.065264 -18.516 < 2e-16 ***
## I(x^2) 0.020854 0.000626 33.311 < 2e-16 ***
## z NA NA NA NA
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.665 on 97 degrees of freedom
## Multiple R-squared: 0.9774, Adjusted R-squared: 0.9769
## F-statistic: 2098 on 2 and 97 DF, p-value: < 2.2e-16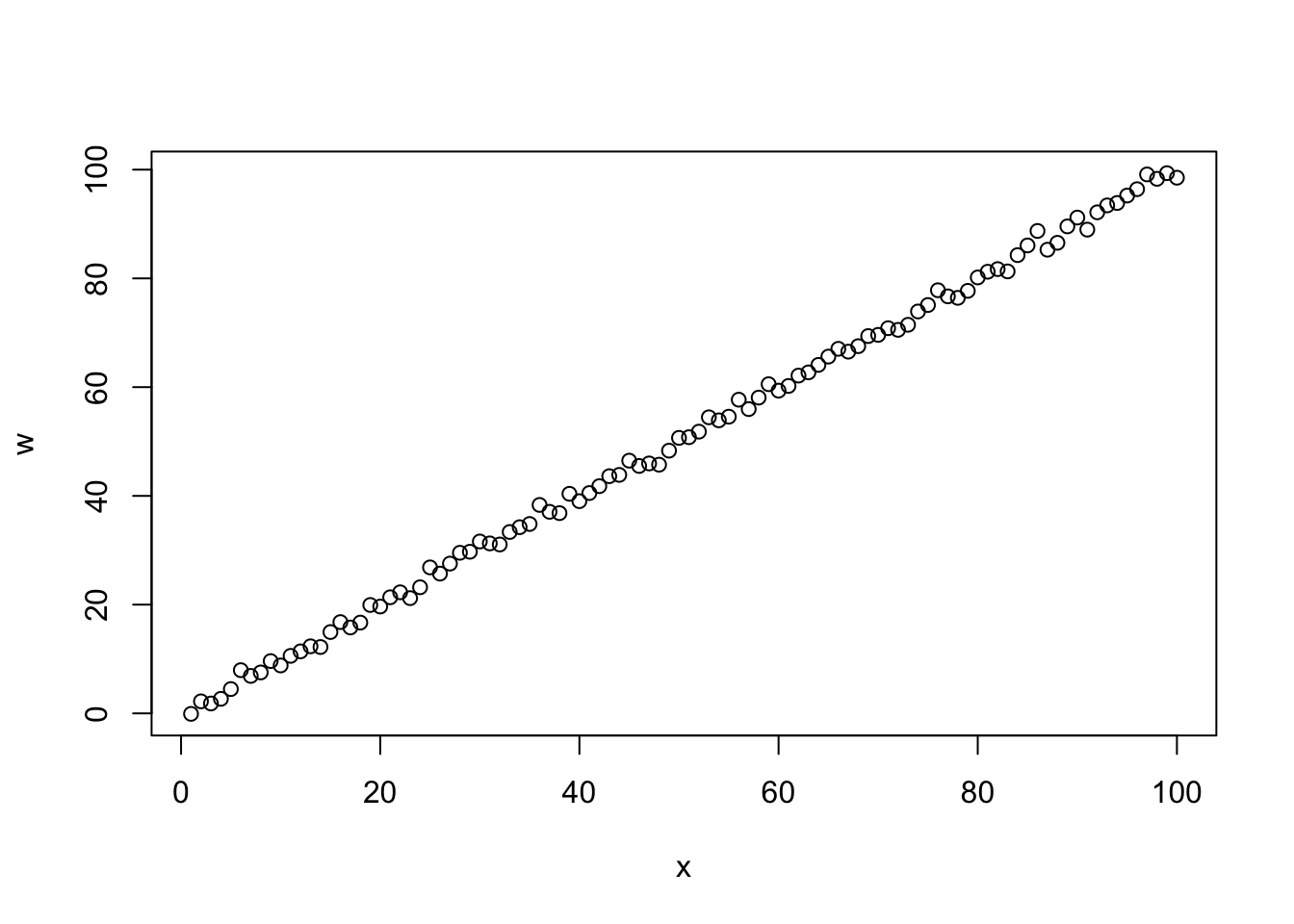
## [1] 0.9993595##
## Call:
## lm(formula = y ~ x + I(x^2) + w)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.5002 -3.2165 -0.2137 2.9738 10.7425
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 11.5987222 1.4380446 8.066 2.06e-12 ***
## x -1.6267839 0.4624638 -3.518 0.000667 ***
## I(x^2) 0.0208978 0.0006284 33.256 < 2e-16 ***
## w 0.4130417 0.4520250 0.914 0.363133
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.669 on 96 degrees of freedom
## Multiple R-squared: 0.9776, Adjusted R-squared: 0.9769
## F-statistic: 1397 on 3 and 96 DF, p-value: < 2.2e-164.2.3 NORMALITY OF ERROR TERMS
Normal Data
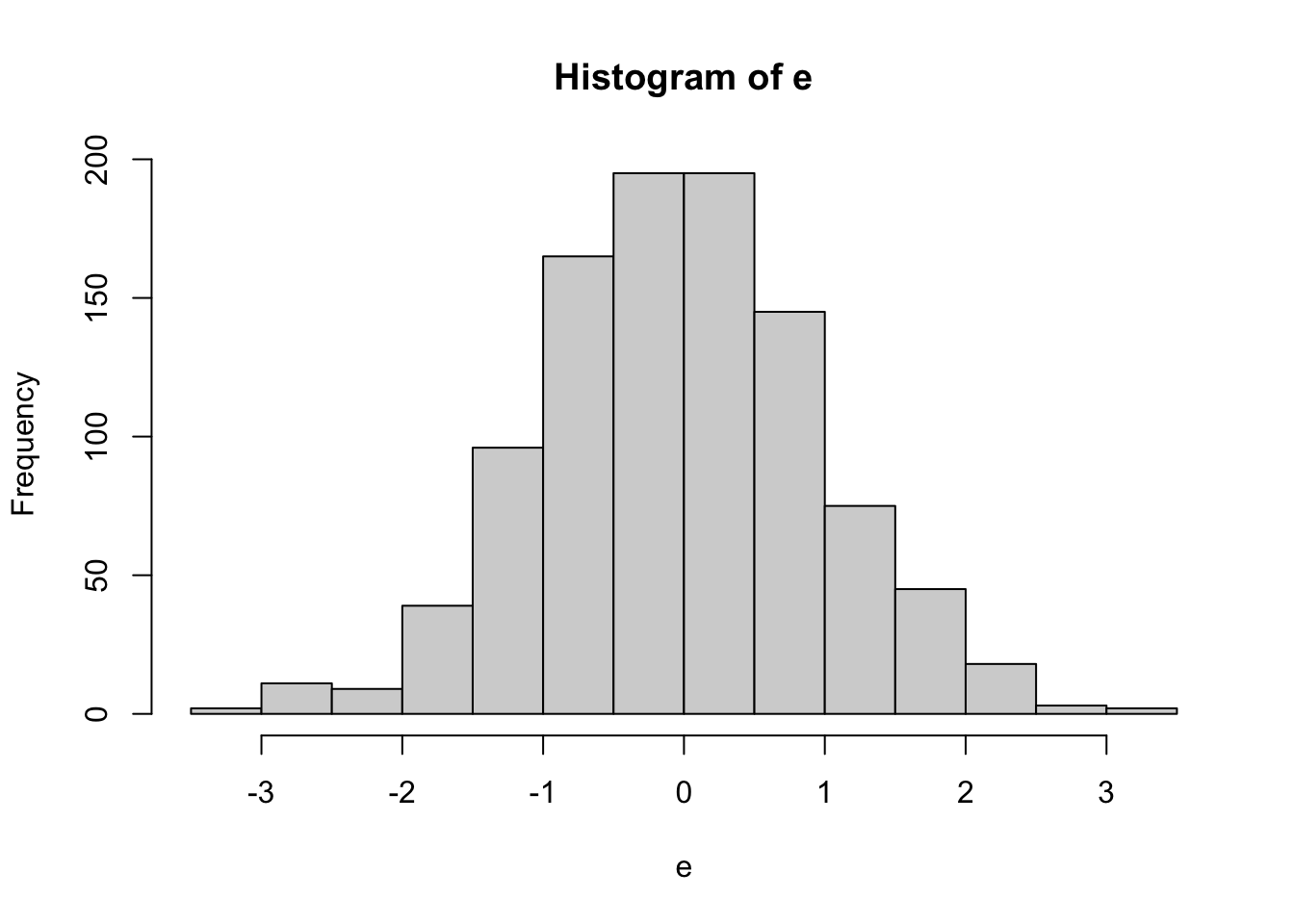
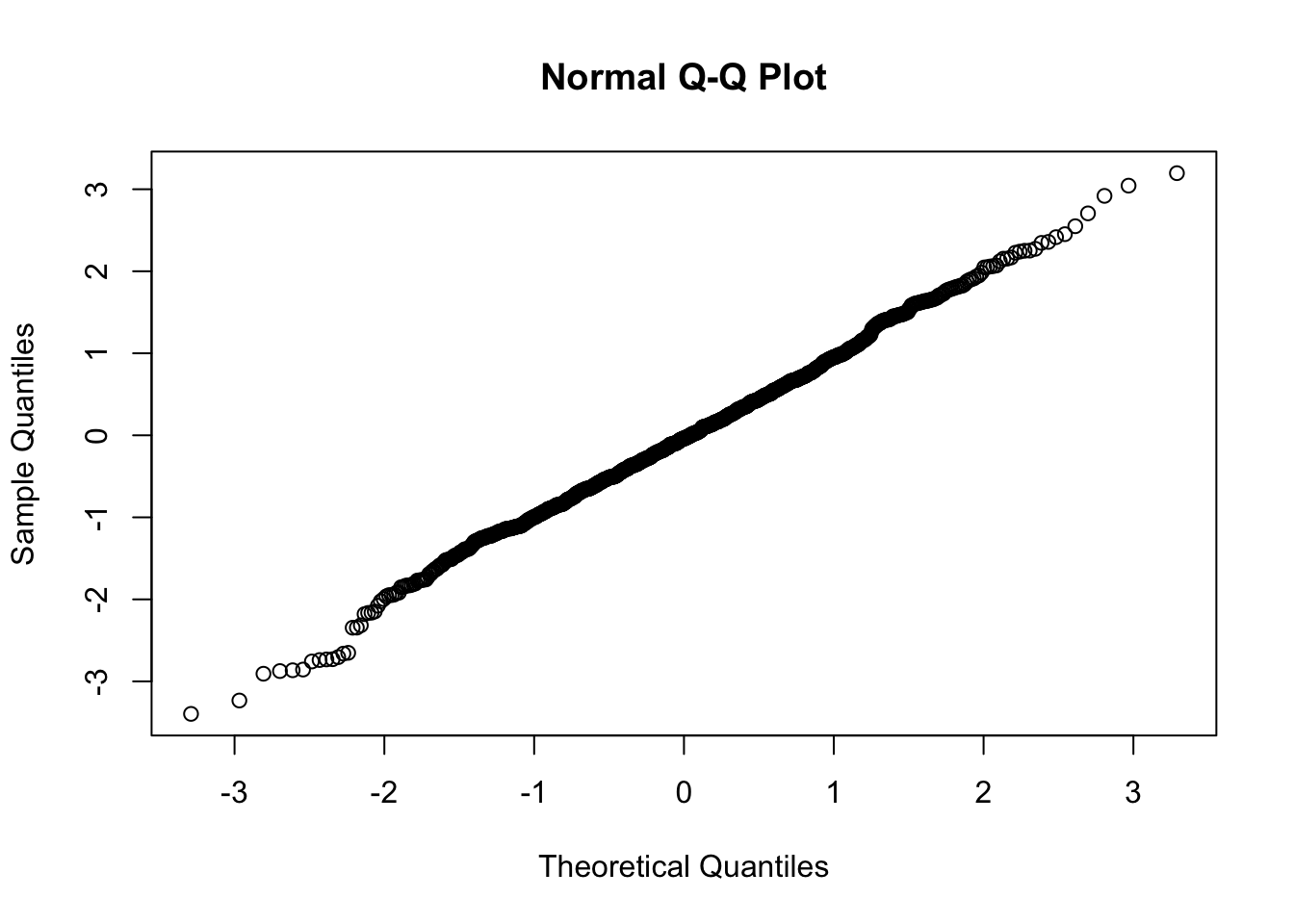
##
## Shapiro-Wilk normality test
##
## data: e
## W = 0.99737, p-value = 0.1053

##
## Shapiro-Wilk normality test
##
## data: e
## W = 0.95765, p-value < 2.2e-16#Model with Nonnormal Errors
x=1:100
y=(200+3*x+20*rnorm(100))^3.56
plot(x,y)
model=lm(y~x)
abline(model,col="red")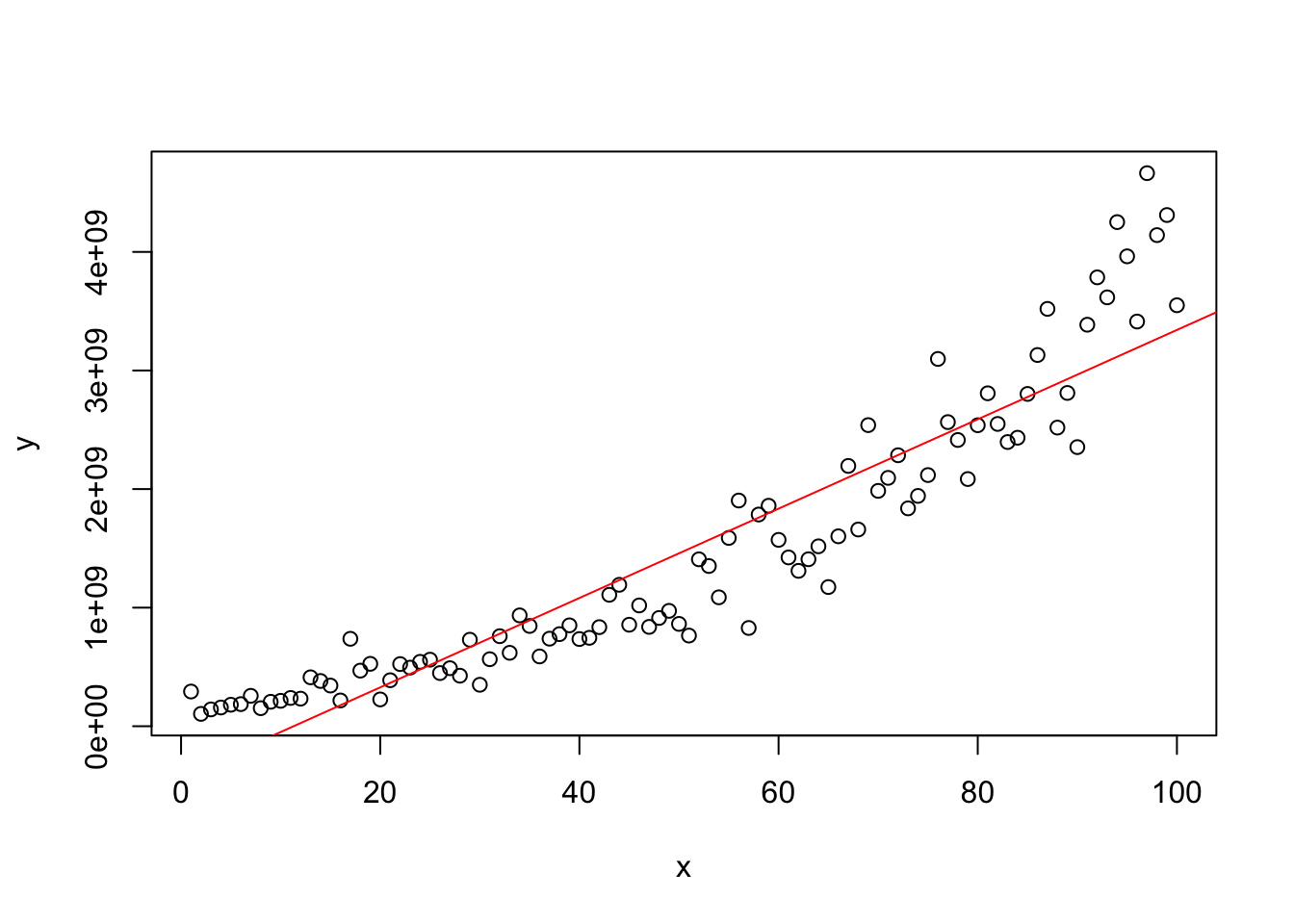
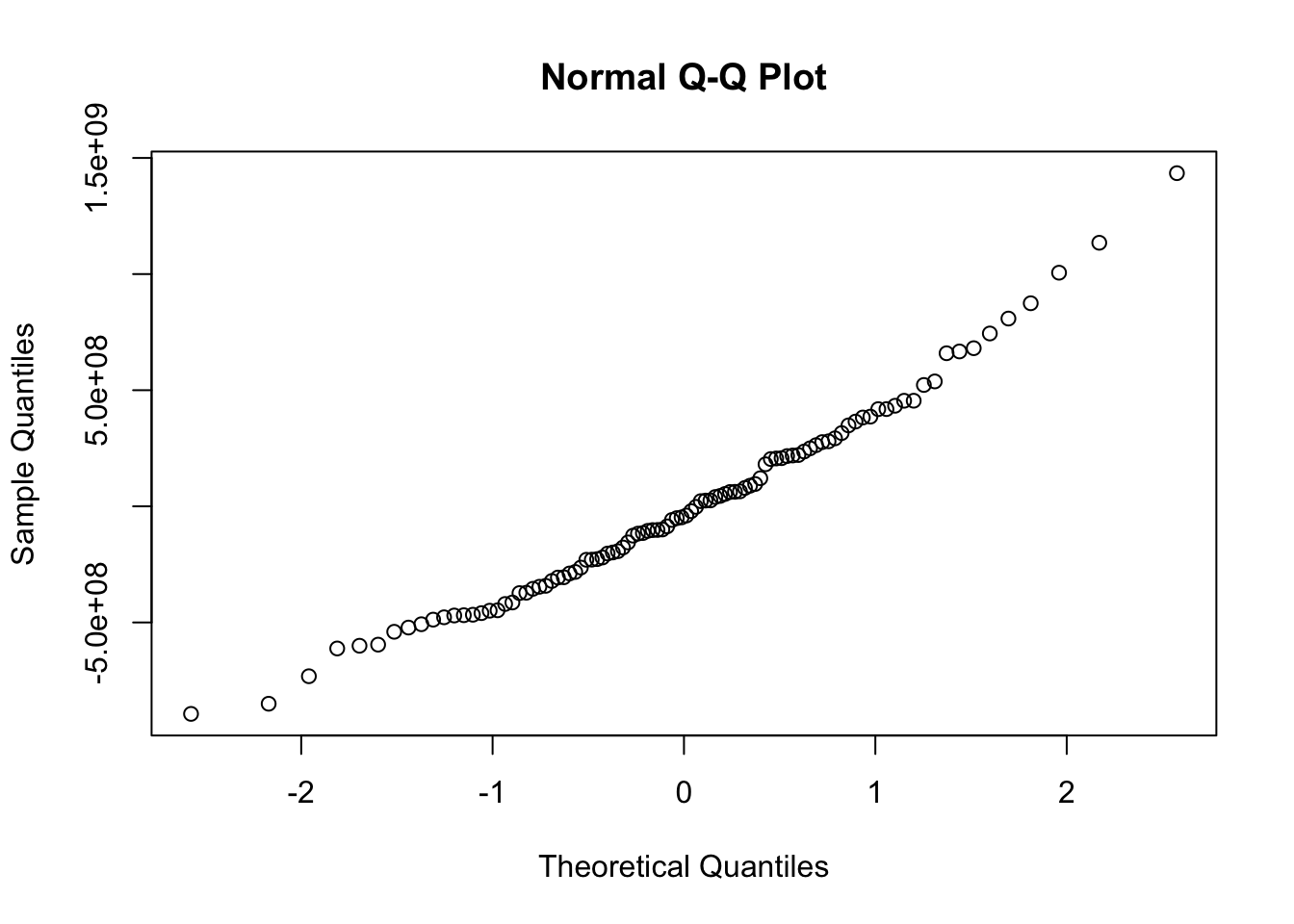
##
## Shapiro-Wilk normality test
##
## data: e
## W = 0.9762, p-value = 0.06698Achieving Normality with Box-Cox Transformations
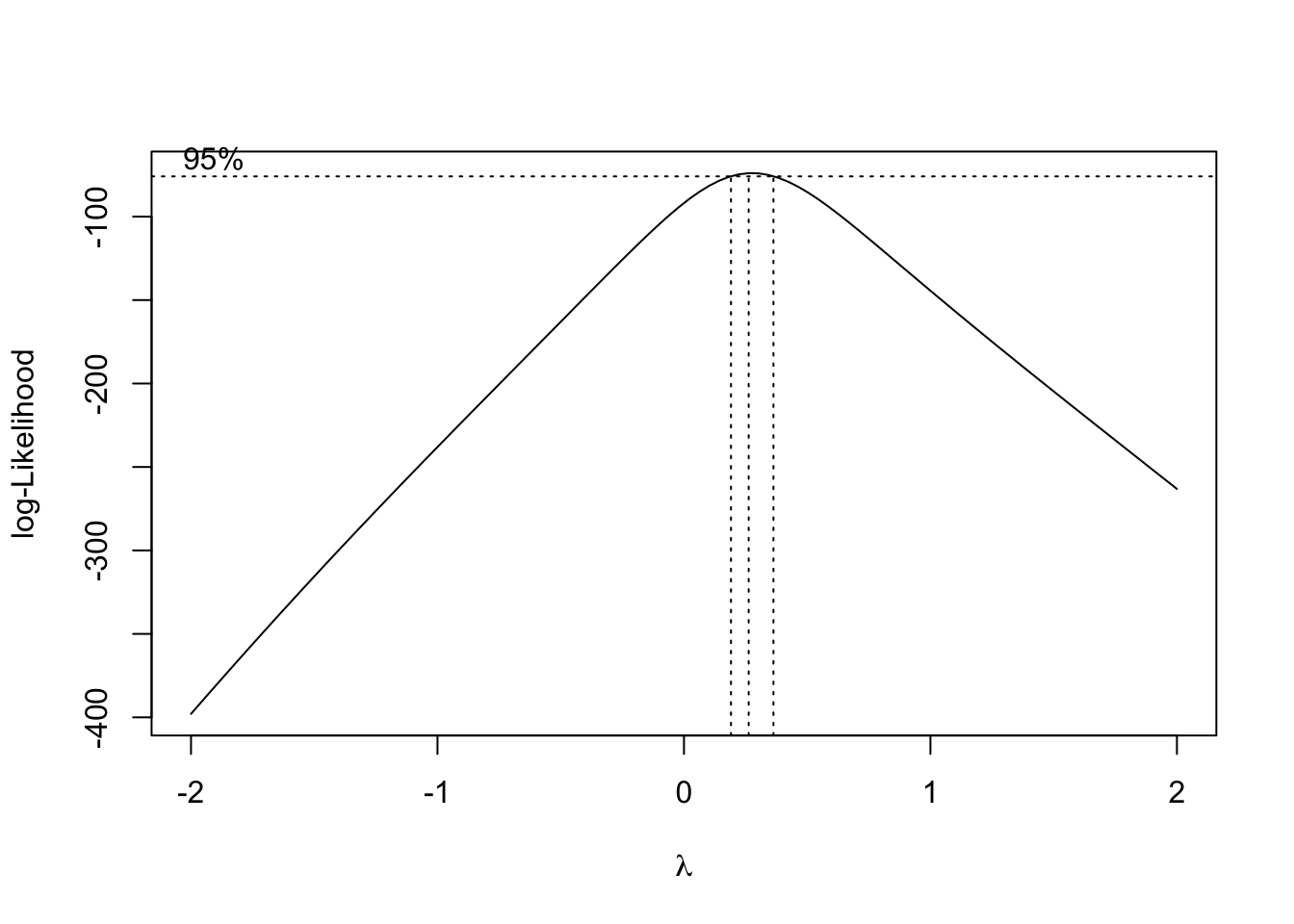
## [,1] [,2]
## [1,] -2.00000000 -397.92136
## [2,] -1.95959596 -391.07290
## [3,] -1.91919192 -384.25863
## [4,] -1.87878788 -377.47860
## [5,] -1.83838384 -370.73285
## [6,] -1.79797980 -364.02145
## [7,] -1.75757576 -357.34444
## [8,] -1.71717172 -350.70173
## [9,] -1.67676768 -344.09323
## [10,] -1.63636364 -337.51874
## [11,] -1.59595960 -330.97801
## [12,] -1.55555556 -324.47075
## [13,] -1.51515152 -317.99653
## [14,] -1.47474747 -311.55492
## [15,] -1.43434343 -305.14532
## [16,] -1.39393939 -298.76710
## [17,] -1.35353535 -292.41952
## [18,] -1.31313131 -286.10171
## [19,] -1.27272727 -279.81275
## [20,] -1.23232323 -273.55156
## [21,] -1.19191919 -267.31696
## [22,] -1.15151515 -261.10768
## [23,] -1.11111111 -254.92228
## [24,] -1.07070707 -248.75925
## [25,] -1.03030303 -242.61694
## [26,] -0.98989899 -236.49355
## [27,] -0.94949495 -230.38725
## [28,] -0.90909091 -224.29600
## [29,] -0.86868687 -218.21775
## [30,] -0.82828283 -212.15037
## [31,] -0.78787879 -206.09166
## [32,] -0.74747475 -200.03947
## [33,] -0.70707071 -193.99168
## [34,] -0.66666667 -187.94628
## [35,] -0.62626263 -181.90162
## [36,] -0.58585859 -175.85624
## [37,] -0.54545455 -169.80932
## [38,] -0.50505051 -163.76098
## [39,] -0.46464646 -157.71182
## [40,] -0.42424242 -151.66482
## [41,] -0.38383838 -145.62388
## [42,] -0.34343434 -139.59592
## [43,] -0.30303030 -133.59176
## [44,] -0.26262626 -127.62421
## [45,] -0.22222222 -121.71491
## [46,] -0.18181818 -115.88857
## [47,] -0.14141414 -110.17968
## [48,] -0.10101010 -104.63395
## [49,] -0.06060606 -99.30216
## [50,] -0.02020202 -94.25458
## [51,] 0.02020202 -89.56714
## [52,] 0.06060606 -85.32617
## [53,] 0.10101010 -81.62837
## [54,] 0.14141414 -78.56756
## [55,] 0.18181818 -76.22513
## [56,] 0.22222222 -74.67758
## [57,] 0.26262626 -73.96405
## [58,] 0.30303030 -74.09086
## [59,] 0.34343434 -75.04712
## [60,] 0.38383838 -76.77031
## [61,] 0.42424242 -79.18669
## [62,] 0.46464646 -82.20836
## [63,] 0.50505051 -85.73659
## [64,] 0.54545455 -89.67670
## [65,] 0.58585859 -93.94605
## [66,] 0.62626263 -98.46543
## [67,] 0.66666667 -103.17232
## [68,] 0.70707071 -108.01550
## [69,] 0.74747475 -112.95015
## [70,] 0.78787879 -117.94625
## [71,] 0.82828283 -122.97679
## [72,] 0.86868687 -128.02325
## [73,] 0.90909091 -133.07213
## [74,] 0.94949495 -138.11241
## [75,] 0.98989899 -143.13797
## [76,] 1.03030303 -148.14365
## [77,] 1.07070707 -153.12676
## [78,] 1.11111111 -158.08593
## [79,] 1.15151515 -163.02045
## [80,] 1.19191919 -167.93077
## [81,] 1.23232323 -172.81755
## [82,] 1.27272727 -177.68197
## [83,] 1.31313131 -182.52546
## [84,] 1.35353535 -187.34951
## [85,] 1.39393939 -192.15579
## [86,] 1.43434343 -196.94595
## [87,] 1.47474747 -201.72166
## [88,] 1.51515152 -206.48454
## [89,] 1.55555556 -211.23618
## [90,] 1.59595960 -215.97808
## [91,] 1.63636364 -220.71171
## [92,] 1.67676768 -225.43839
## [93,] 1.71717172 -230.15943
## [94,] 1.75757576 -234.87603
## [95,] 1.79797980 -239.58931
## [96,] 1.83838384 -244.30033
## [97,] 1.87878788 -249.01002
## [98,] 1.91919192 -253.71923
## [99,] 1.95959596 -258.42886
## [100,] 2.00000000 -263.13980## [1] 57lambda=boxcox.results$x[which.max(boxcox.results$y)]
y.tilde=y^lambda
trans.model=lm(y.tilde~x)
plot(x,y.tilde)
abline(trans.model,col="red")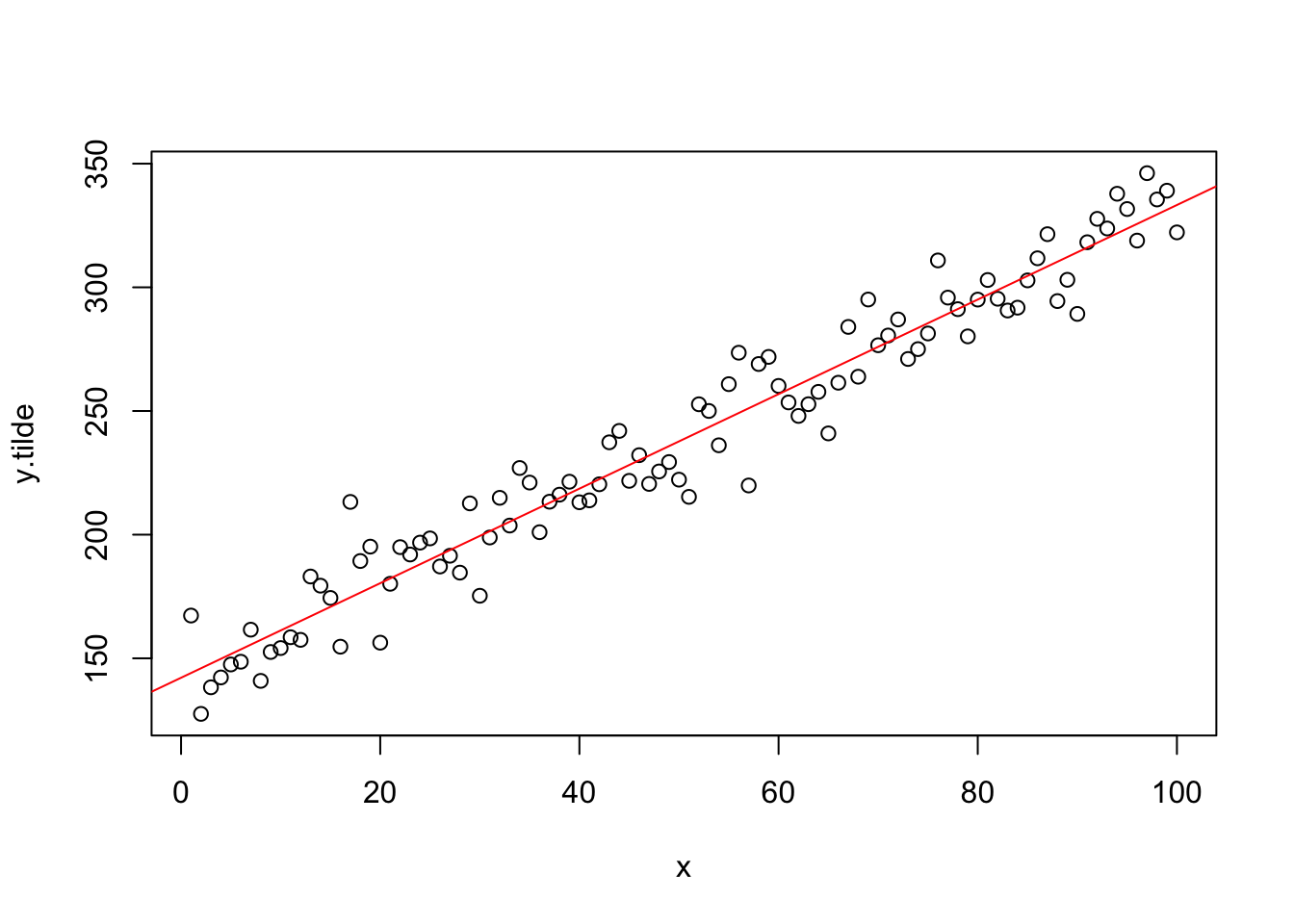
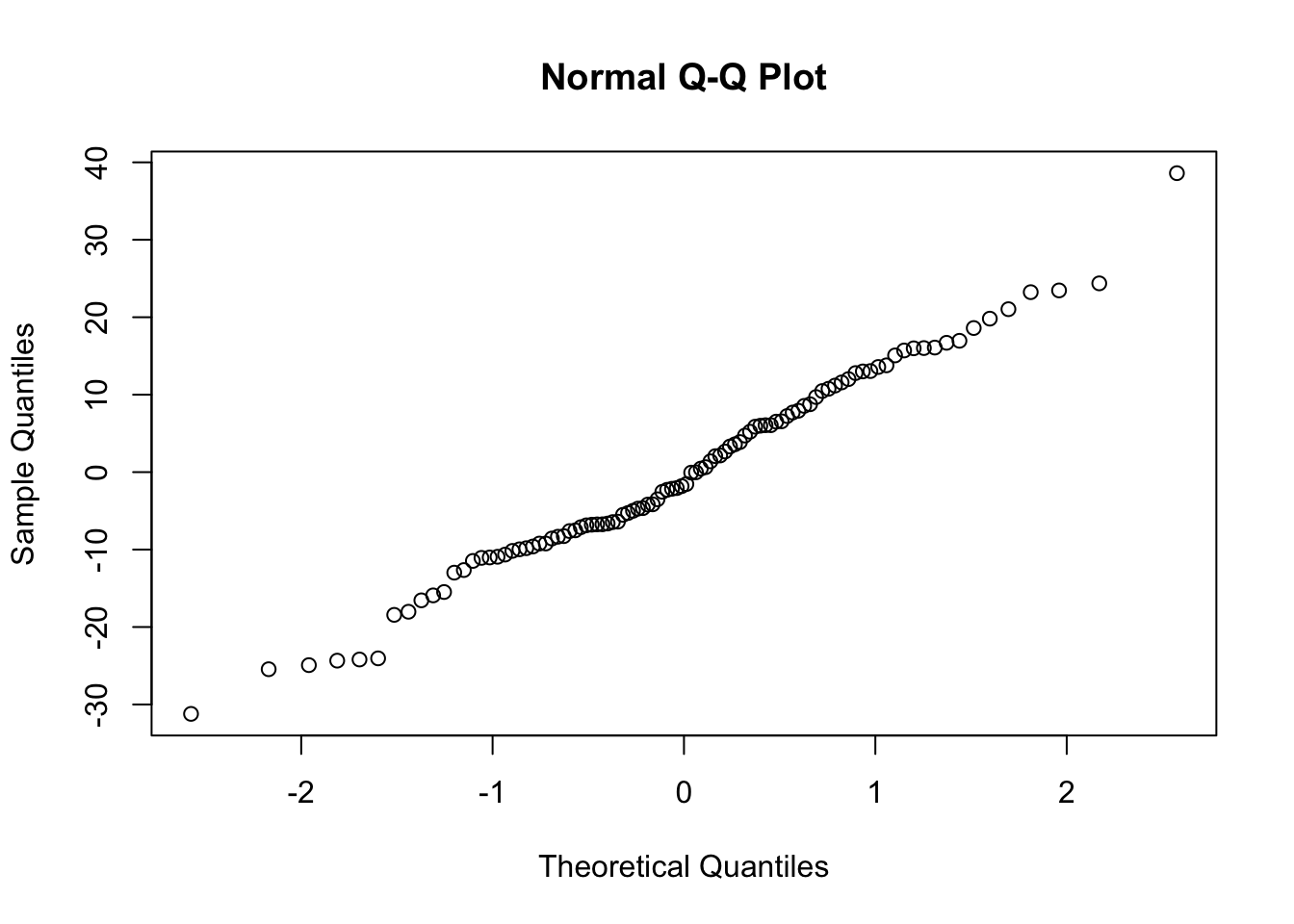
##
## Shapiro-Wilk normality test
##
## data: e.tilde
## W = 0.98912, p-value = 0.594Using Transformed Model to Make Predictions
new.data=data.frame(x=runif(10,min=0,max=100))
y.tilde.hat=predict(trans.model,newdata=new.data)
plot(new.data$x,y.tilde.hat)
abline(trans.model,col="red")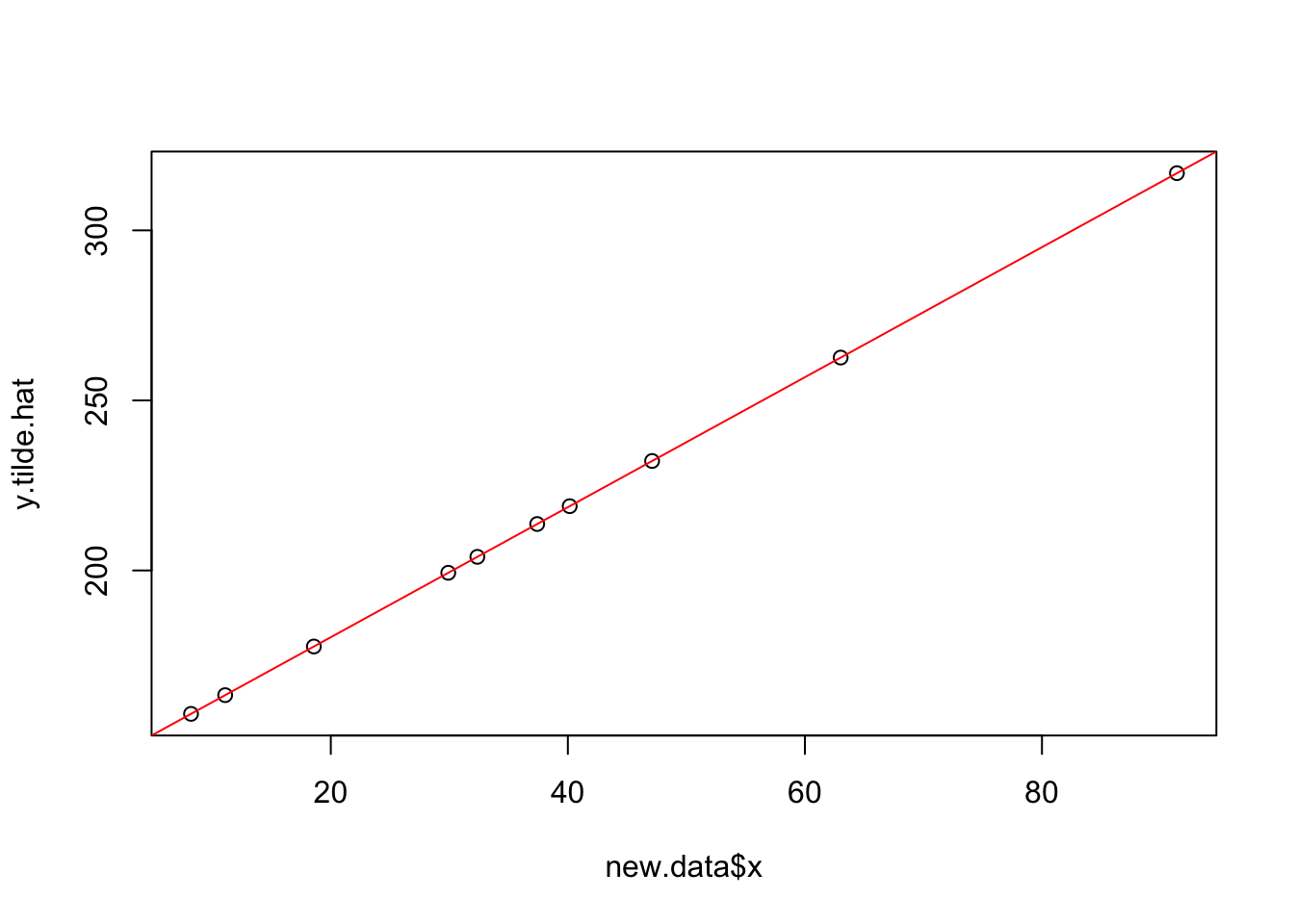
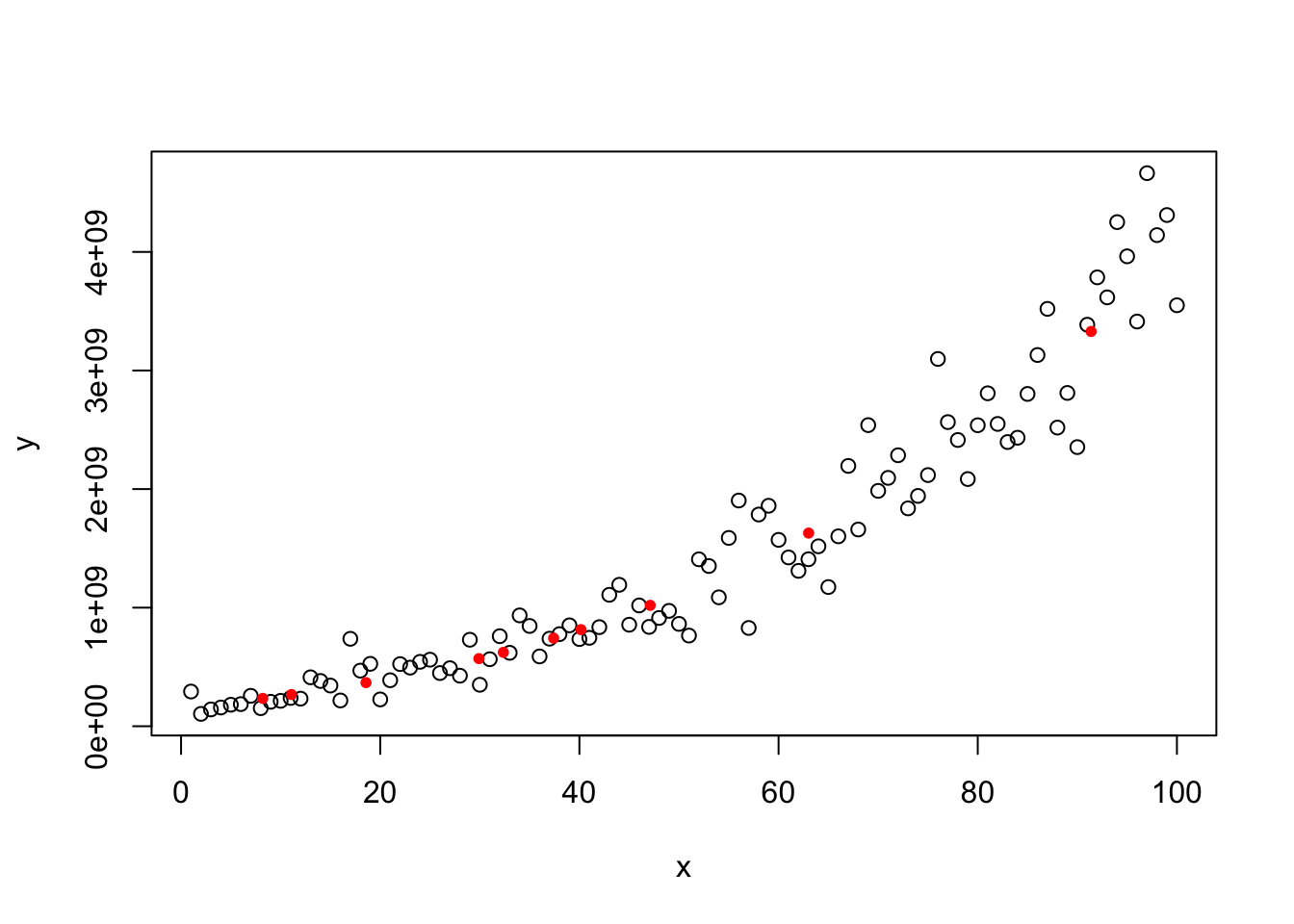
4.2.4 Constancy of Error Variance (Homoscedasticity)
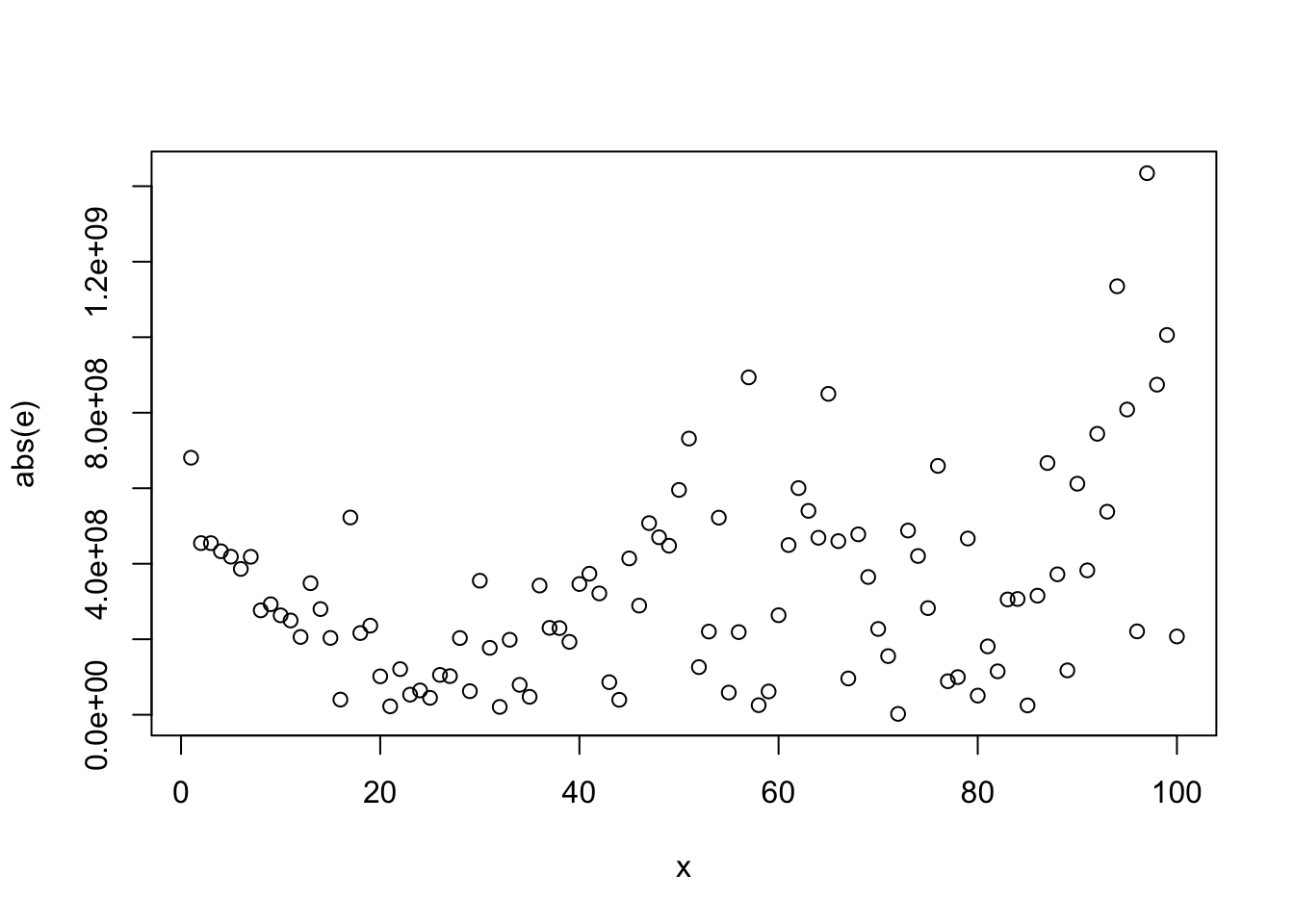
##
## Modified robust Brown-Forsythe Levene-type test based on the absolute deviations from
## the median
##
## data: e
## Test Statistic = 7.3656, p-value = 0.007856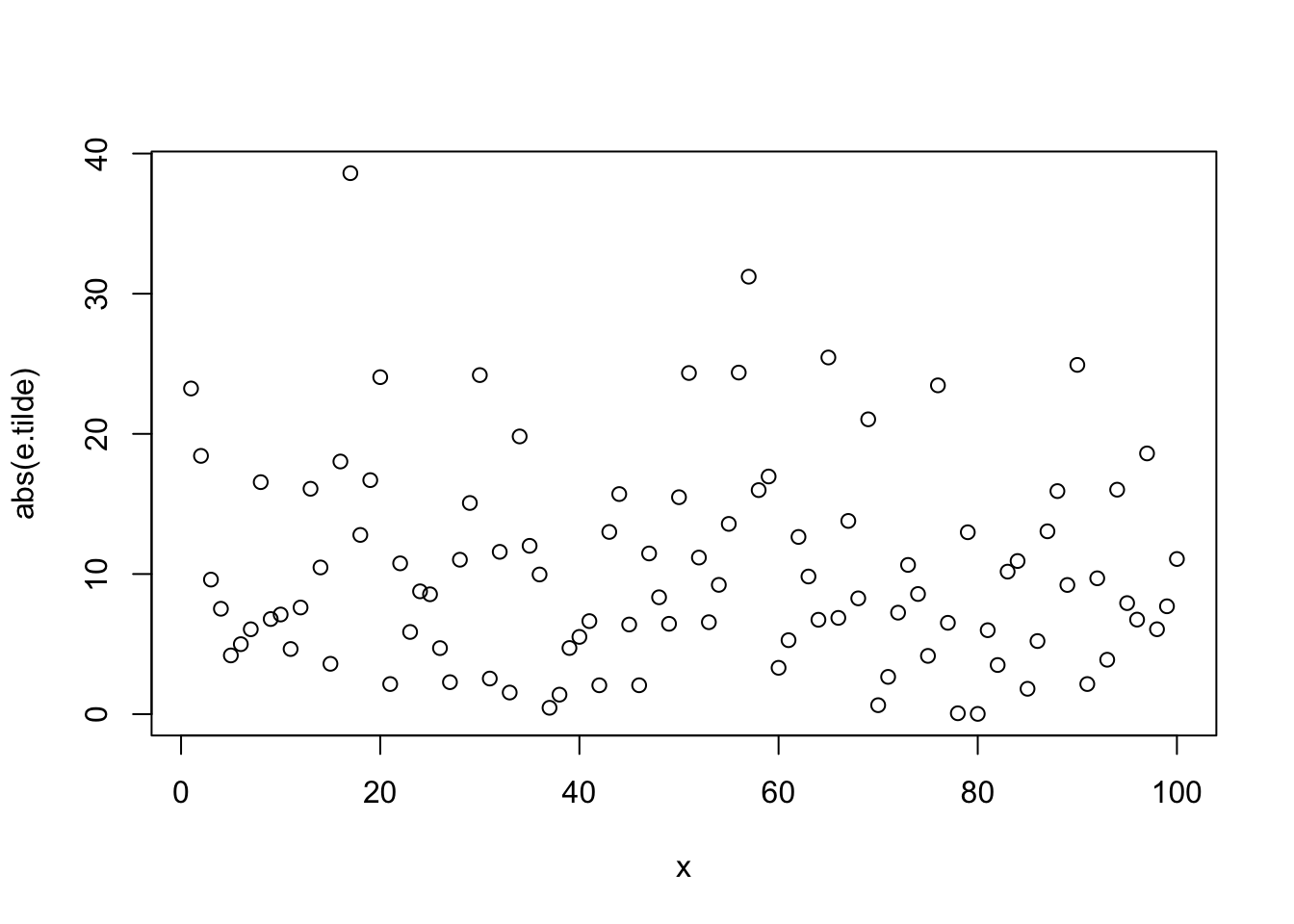
##
## Modified robust Brown-Forsythe Levene-type test based on the absolute deviations from
## the median
##
## data: e.tilde
## Test Statistic = 0.24102, p-value = 0.6246A Permutation Test Example
Creating Data
set.seed(403)
x1=runif(100,min=0,max=100)
x2=runif(100,min=0,max=100)
epsilon=runif(100,min=-50,max=50)
y=200+3*x1+5*x2+epsilon
mydata=data.frame(y,x1,x2)
plot(mydata)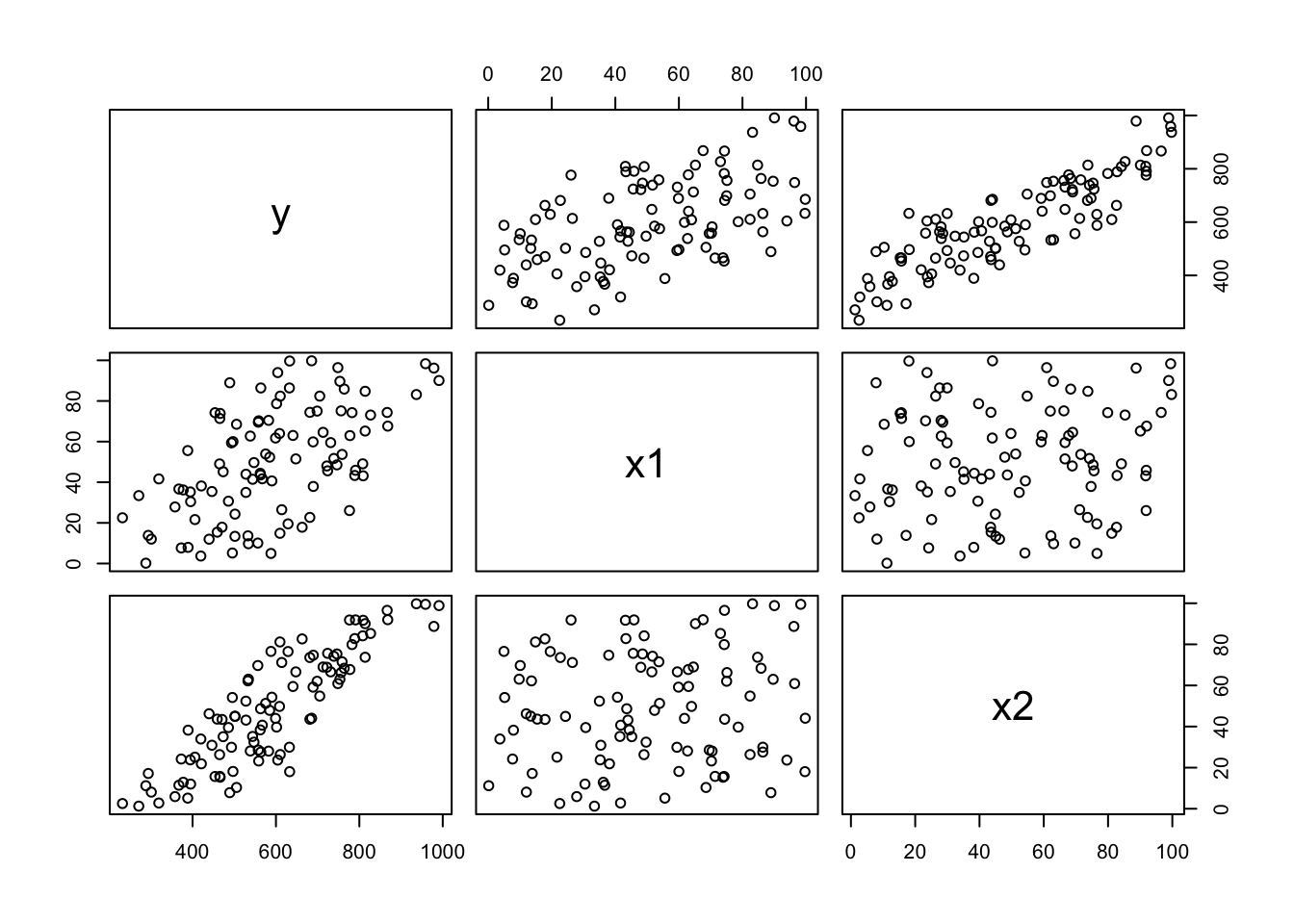
##
## Call:
## lm(formula = y ~ x1 + x2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -51.954 -25.658 -3.725 26.312 54.757
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 206.5557 7.7986 26.49 <2e-16 ***
## x1 2.8750 0.1112 25.85 <2e-16 ***
## x2 4.9691 0.1091 45.53 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 29.47 on 97 degrees of freedom
## Multiple R-squared: 0.9695, Adjusted R-squared: 0.9689
## F-statistic: 1543 on 2 and 97 DF, p-value: < 2.2e-16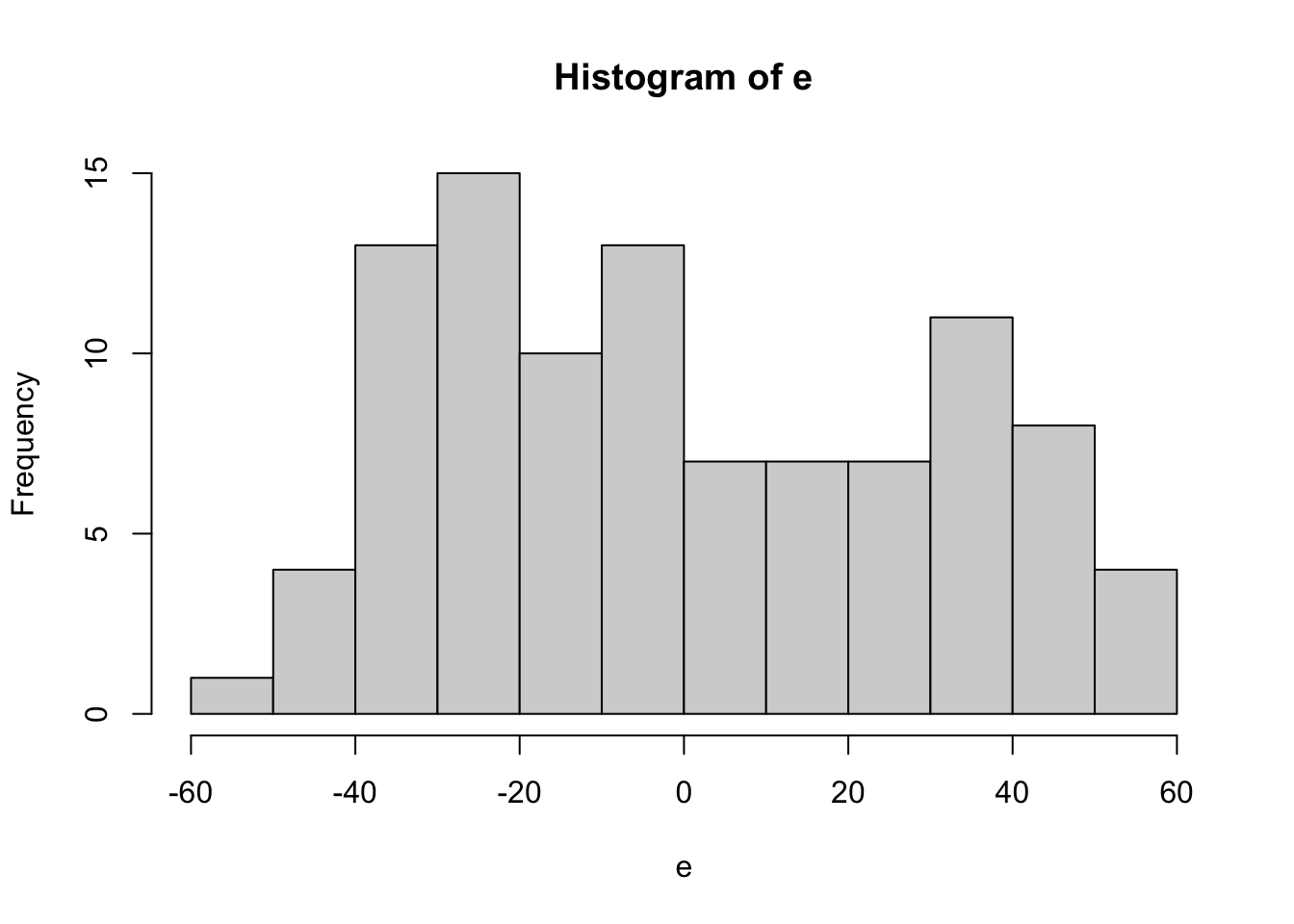

##
## Shapiro-Wilk normality test
##
## data: e
## W = 0.94432, p-value = 0.000357Model is y = beta0 + beta1*x1 + beta2*x2
Permutation Test for H0: beta2=0
N=1000
beta2.star.vect = 1:N
for(i in 1:N){
temp.x2 = sample(x2,
size=length(x2),
replace=FALSE)
temp.model=lm(y~x1+temp.x2)
temp.betahat=coef(temp.model)
beta2.star.vect[i]=temp.betahat[3]
}
hist(beta2.star.vect) #All beta2 values from permutation test are stored in this vector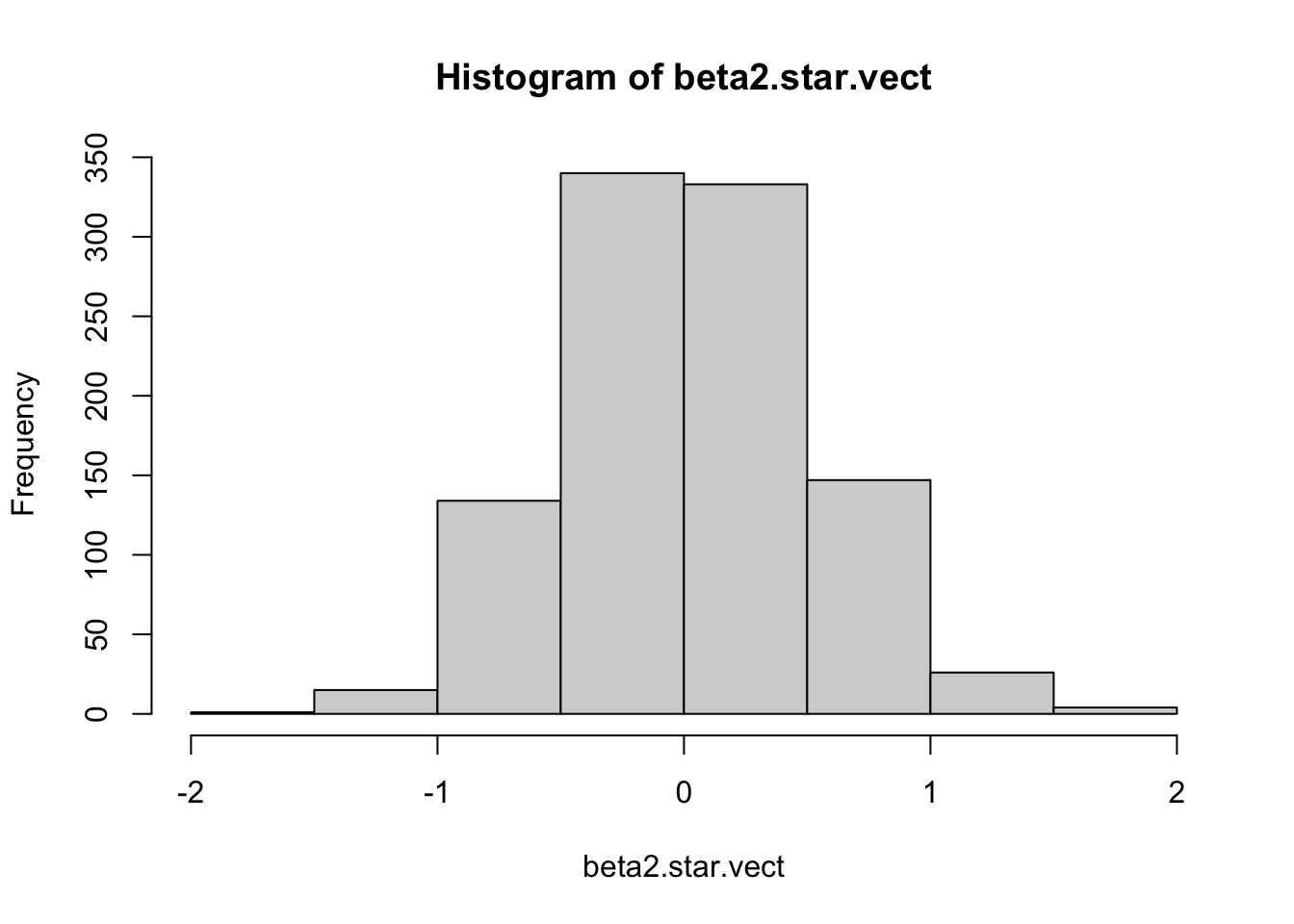
## x2
## 4.969105How do they compare?
## [1] -1.800433 1.622146## [1] 0